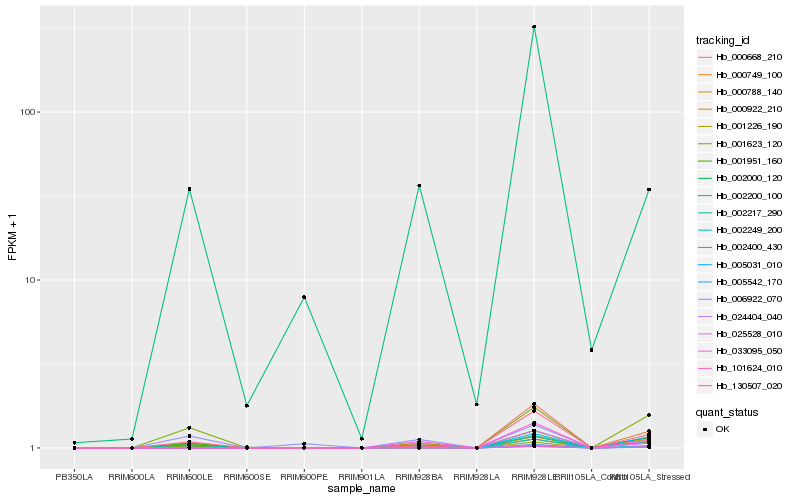
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_210 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L12, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002217_290 |
0.0028416565 |
- |
- |
hypothetical protein JCGZ_19038 [Jatropha curcas] |
| 3 |
Hb_130507_020 |
0.1769744756 |
- |
- |
- |
| 4 |
Hb_000668_210 |
0.2019465404 |
- |
- |
hypothetical protein PHAVU_006G031500g [Phaseolus vulgaris] |
| 5 |
Hb_001226_190 |
0.2071211948 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 6 |
Hb_001951_160 |
0.2390082695 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 7 |
Hb_024404_040 |
0.2395451238 |
- |
- |
hypothetical protein JCGZ_22351 [Jatropha curcas] |
| 8 |
Hb_101624_010 |
0.2495670831 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 9 |
Hb_005031_010 |
0.2495730594 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 10 |
Hb_000749_100 |
0.2517411883 |
- |
- |
- |
| 11 |
Hb_002000_120 |
0.2567105886 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_001623_120 |
0.2576568975 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X2 [Jatropha curcas] |
| 13 |
Hb_006922_070 |
0.2633906968 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 14 |
Hb_005542_170 |
0.2694974892 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_002200_100 |
0.2699639243 |
- |
- |
PREDICTED: pathogen-related protein-like [Jatropha curcas] |
| 16 |
Hb_000788_140 |
0.2727976623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_033095_050 |
0.2743310262 |
- |
- |
hypothetical protein VITISV_038692 [Vitis vinifera] |
| 18 |
Hb_002400_430 |
0.2767738638 |
- |
- |
- |
| 19 |
Hb_025528_010 |
0.2849391123 |
- |
- |
PREDICTED: uncharacterized protein LOC102596706 [Solanum tuberosum] |
| 20 |
Hb_002249_200 |
0.2902248635 |
transcription factor |
TF Family: LFY |
LEAFY-like protein [Pistacia chinensis] |