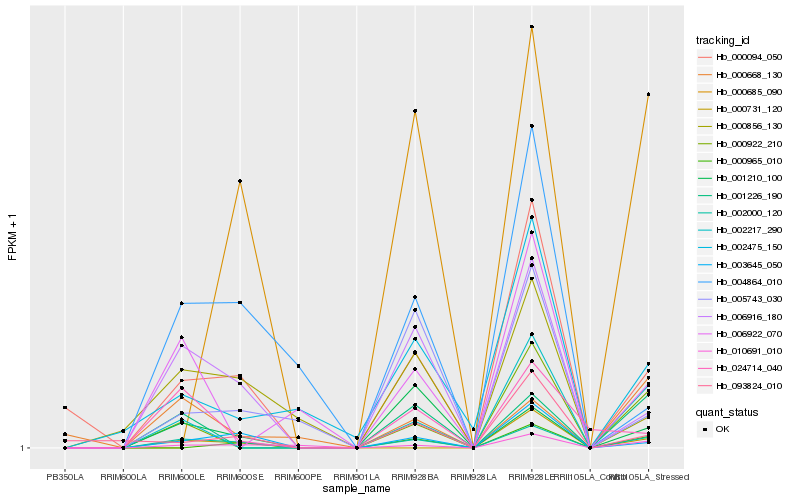
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002000_120 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_006916_180 |
0.1781454805 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 3 |
Hb_000094_050 |
0.2022561813 |
- |
- |
- |
| 4 |
Hb_003645_050 |
0.2182872949 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_010691_010 |
0.2370130324 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 6 |
Hb_001210_100 |
0.2459239074 |
- |
- |
Gibberellin 20 oxidase, putative [Ricinus communis] |
| 7 |
Hb_002217_290 |
0.2560649018 |
- |
- |
hypothetical protein JCGZ_19038 [Jatropha curcas] |
| 8 |
Hb_000922_210 |
0.2567105886 |
- |
- |
PREDICTED: 50S ribosomal protein L12, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001226_190 |
0.2701056133 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 10 |
Hb_000856_130 |
0.2729451474 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_005743_030 |
0.2886704001 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 12 |
Hb_000965_010 |
0.2892579471 |
- |
- |
hypothetical protein VITISV_016917 [Vitis vinifera] |
| 13 |
Hb_004864_010 |
0.2896193862 |
- |
- |
hypothetical protein CICLE_v10003960mg, partial [Citrus clementina] |
| 14 |
Hb_002475_150 |
0.2940403404 |
- |
- |
PREDICTED: importin subunit beta-3-like [Jatropha curcas] |
| 15 |
Hb_024714_040 |
0.3019738783 |
- |
- |
PREDICTED: DNA mismatch repair protein PMS1 [Jatropha curcas] |
| 16 |
Hb_000668_130 |
0.3077792991 |
- |
- |
- |
| 17 |
Hb_006922_070 |
0.3125169467 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 18 |
Hb_000731_120 |
0.3145892802 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 19 |
Hb_000685_090 |
0.3163633769 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 20 |
Hb_093824_010 |
0.31788272 |
- |
- |
PREDICTED: uncharacterized protein LOC103326132 [Prunus mume] |