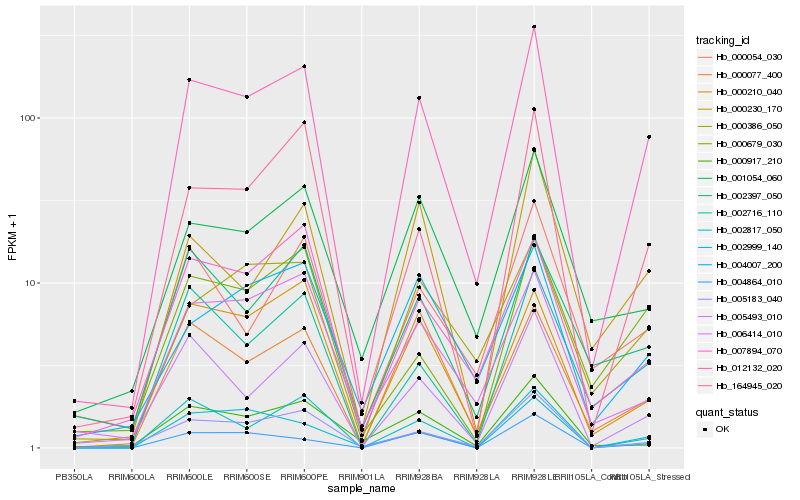
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012132_020 |
0.0 |
- |
- |
glutamine synthetase [Hevea brasiliensis] |
| 2 |
Hb_005183_040 |
0.1379308718 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 3 |
Hb_000210_040 |
0.148030658 |
- |
- |
PREDICTED: putative HVA22-like protein g [Jatropha curcas] |
| 4 |
Hb_164945_020 |
0.1481155907 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 5 |
Hb_005493_010 |
0.1500611648 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_002716_110 |
0.1558717215 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 7 |
Hb_002817_050 |
0.1560962971 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0004s16810g, partial [Populus trichocarpa] |
| 8 |
Hb_000386_050 |
0.1572233511 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000077_400 |
0.1581536052 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_006414_010 |
0.1612672532 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 11 |
Hb_007894_070 |
0.1631236869 |
- |
- |
PREDICTED: uncharacterized protein LOC105632955 [Jatropha curcas] |
| 12 |
Hb_004007_200 |
0.1694330995 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY5 [Jatropha curcas] |
| 13 |
Hb_001054_060 |
0.1700579786 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 14 |
Hb_000230_170 |
0.1702091321 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 15 |
Hb_002999_140 |
0.1704766616 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 16 |
Hb_004864_010 |
0.1734651908 |
- |
- |
hypothetical protein CICLE_v10003960mg, partial [Citrus clementina] |
| 17 |
Hb_000054_030 |
0.1736137361 |
- |
- |
hypothetical protein POPTR_0010s17680g [Populus trichocarpa] |
| 18 |
Hb_002397_050 |
0.1737425281 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000679_030 |
0.1747033289 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5 [Jatropha curcas] |
| 20 |
Hb_000917_210 |
0.1787050781 |
- |
- |
conserved hypothetical protein [Ricinus communis] |