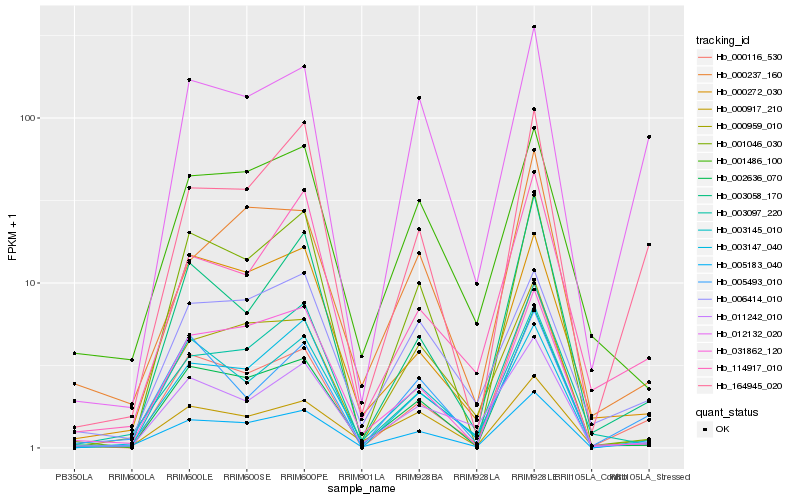
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005183_040 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 2 |
Hb_164945_020 |
0.1248083374 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 3 |
Hb_002636_070 |
0.1288542457 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 4 |
Hb_000917_210 |
0.1326974163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000959_010 |
0.1331817122 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_011242_010 |
0.135232829 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 7 |
Hb_003145_010 |
0.1360413207 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 8 |
Hb_012132_020 |
0.1379308718 |
- |
- |
glutamine synthetase [Hevea brasiliensis] |
| 9 |
Hb_005493_010 |
0.1405079295 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_003147_040 |
0.1409464163 |
- |
- |
PREDICTED: probable calcium-binding protein CML18 [Jatropha curcas] |
| 11 |
Hb_031862_120 |
0.1412981463 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 12 |
Hb_003058_170 |
0.1443963636 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000116_530 |
0.1449391364 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 3; Short=HMG-CoA reductase 3 [Hevea brasiliensis] |
| 14 |
Hb_000237_160 |
0.1462895292 |
- |
- |
BEL1-like homeodomain protein 1 [Morus notabilis] |
| 15 |
Hb_003097_220 |
0.1466910487 |
- |
- |
hypothetical protein POPTR_0001s06150g [Populus trichocarpa] |
| 16 |
Hb_114917_010 |
0.1476102261 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000272_030 |
0.1479818143 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001046_030 |
0.1506059263 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_001486_100 |
0.1507998482 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 20 |
Hb_006414_010 |
0.1511207112 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |