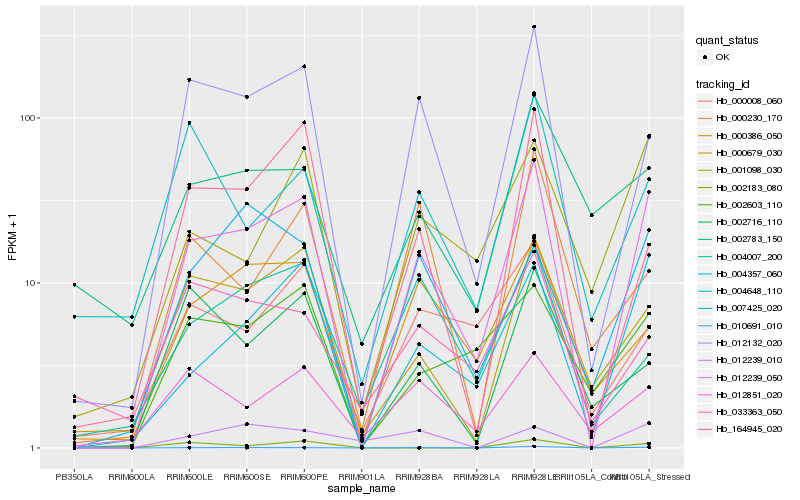
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012239_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004648_110 |
0.1699409368 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_010691_010 |
0.1762796729 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 4 |
Hb_012132_020 |
0.1817748887 |
- |
- |
glutamine synthetase [Hevea brasiliensis] |
| 5 |
Hb_001098_030 |
0.2132719481 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Populus euphratica] |
| 6 |
Hb_000679_030 |
0.214315198 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5 [Jatropha curcas] |
| 7 |
Hb_002603_110 |
0.2194672081 |
- |
- |
PREDICTED: uncharacterized protein LOC105646325 [Jatropha curcas] |
| 8 |
Hb_000386_050 |
0.2280917113 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_164945_020 |
0.2296775447 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 10 |
Hb_012851_020 |
0.2324434975 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_000008_060 |
0.2348797575 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_007425_020 |
0.2381984915 |
transcription factor |
TF Family: ERF |
PREDICTED: pathogenesis-related genes transcriptional activator PTI6-like [Jatropha curcas] |
| 13 |
Hb_012239_010 |
0.2504586813 |
- |
- |
- |
| 14 |
Hb_002783_150 |
0.2542110007 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 15 |
Hb_002716_110 |
0.2553277767 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 16 |
Hb_004357_060 |
0.2578005346 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 17 |
Hb_000230_170 |
0.2587177402 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 18 |
Hb_004007_200 |
0.2606551794 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY5 [Jatropha curcas] |
| 19 |
Hb_033363_050 |
0.2631542139 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002183_080 |
0.2633147718 |
- |
- |
catalytic, putative [Ricinus communis] |