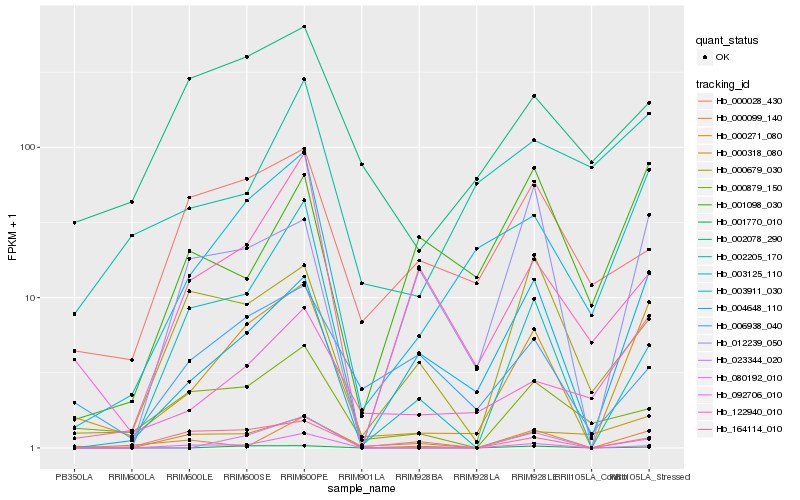
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000271_080 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s08460g [Populus trichocarpa] |
| 2 |
Hb_003911_030 |
0.2070467307 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_004648_110 |
0.237360404 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_023344_020 |
0.2686817842 |
- |
- |
- |
| 5 |
Hb_001770_010 |
0.2850370308 |
- |
- |
hypothetical protein JCGZ_15779 [Jatropha curcas] |
| 6 |
Hb_000318_080 |
0.2875378076 |
- |
- |
PREDICTED: la-related protein 6C [Jatropha curcas] |
| 7 |
Hb_092706_010 |
0.2908607936 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 8 |
Hb_000879_150 |
0.2953377571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002078_290 |
0.2960625643 |
- |
- |
PREDICTED: uncharacterized protein LOC100852520 [Vitis vinifera] |
| 10 |
Hb_080192_010 |
0.2999307789 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 11 |
Hb_164114_010 |
0.3030149731 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_000099_140 |
0.3065243023 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 13 |
Hb_012239_050 |
0.3067127249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006938_040 |
0.3087268159 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 15 |
Hb_002205_170 |
0.3096907331 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 16 |
Hb_000679_030 |
0.3181120865 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5 [Jatropha curcas] |
| 17 |
Hb_003125_110 |
0.3235486349 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |
| 18 |
Hb_001098_030 |
0.3251762301 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Populus euphratica] |
| 19 |
Hb_122940_010 |
0.3261402444 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Populus euphratica] |
| 20 |
Hb_000028_430 |
0.326325473 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |