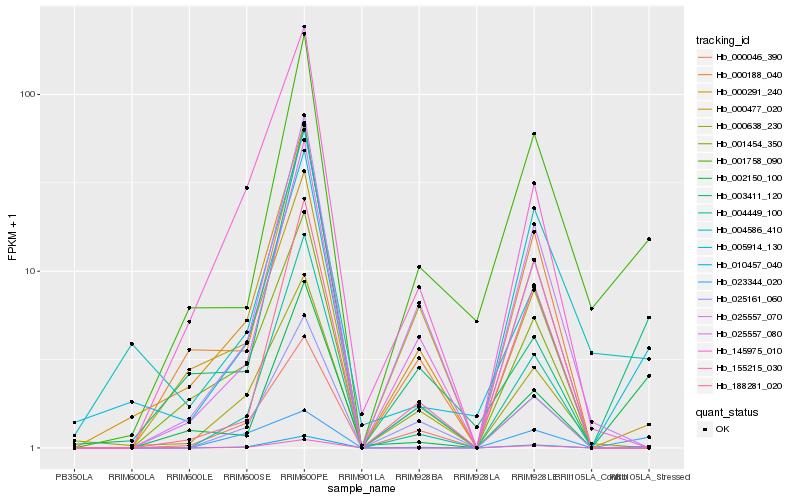
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010457_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC101491058 [Cicer arietinum] |
| 2 |
Hb_005914_130 |
0.1649739824 |
- |
- |
- |
| 3 |
Hb_001758_090 |
0.1825094579 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 4 |
Hb_002150_100 |
0.1957784845 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Jatropha curcas] |
| 5 |
Hb_025161_060 |
0.2075335895 |
- |
- |
hypothetical protein JCGZ_23770 [Jatropha curcas] |
| 6 |
Hb_025557_080 |
0.2138939338 |
transcription factor |
TF Family: zf-HD |
- |
| 7 |
Hb_000046_390 |
0.2147466395 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 8 |
Hb_025557_070 |
0.2180452622 |
transcription factor |
TF Family: zf-HD |
hypothetical protein JCGZ_11626 [Jatropha curcas] |
| 9 |
Hb_004449_100 |
0.2218057433 |
- |
- |
- |
| 10 |
Hb_000291_240 |
0.2261792463 |
- |
- |
hypothetical protein POPTR_0005s16430g [Populus trichocarpa] |
| 11 |
Hb_003411_120 |
0.2264029228 |
- |
- |
hypothetical protein POPTR_0010s13670g [Populus trichocarpa] |
| 12 |
Hb_000638_230 |
0.2280419488 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 13 |
Hb_188281_020 |
0.2287092125 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 14 |
Hb_001454_350 |
0.2323661146 |
- |
- |
PREDICTED: uncharacterized protein LOC105643449 [Jatropha curcas] |
| 15 |
Hb_004586_410 |
0.2340524022 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 16 |
Hb_155215_030 |
0.2343443027 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 17 |
Hb_023344_020 |
0.2345734978 |
- |
- |
- |
| 18 |
Hb_000188_040 |
0.2355638205 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 19 |
Hb_000477_020 |
0.2379682284 |
- |
- |
hypothetical protein EUGRSUZ_C011052, partial [Eucalyptus grandis] |
| 20 |
Hb_145975_010 |
0.239070592 |
- |
- |
cytochrome P450, putative [Ricinus communis] |