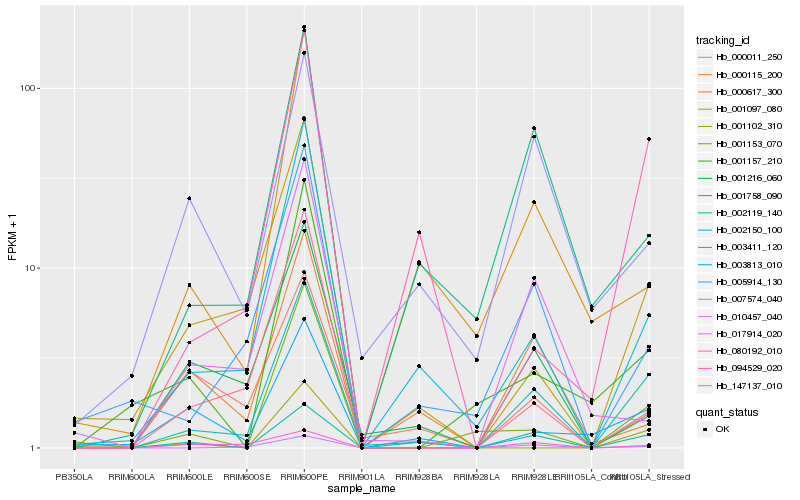
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002150_100 |
0.0 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Jatropha curcas] |
| 2 |
Hb_003411_120 |
0.1886382688 |
- |
- |
hypothetical protein POPTR_0010s13670g [Populus trichocarpa] |
| 3 |
Hb_010457_040 |
0.1957784845 |
- |
- |
PREDICTED: uncharacterized protein LOC101491058 [Cicer arietinum] |
| 4 |
Hb_002119_140 |
0.1973098027 |
- |
- |
PREDICTED: uncharacterized protein LOC105634172 [Jatropha curcas] |
| 5 |
Hb_001097_080 |
0.2153696771 |
transcription factor |
TF Family: MYB-related |
triptychon and cpc, putative [Ricinus communis] |
| 6 |
Hb_005914_130 |
0.2153878851 |
- |
- |
- |
| 7 |
Hb_001758_090 |
0.2248845519 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 8 |
Hb_094529_020 |
0.2317051087 |
- |
- |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 9 |
Hb_001153_070 |
0.2357662115 |
- |
- |
- |
| 10 |
Hb_001216_060 |
0.2412359394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000617_300 |
0.2464038538 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 12 |
Hb_003813_010 |
0.2475208658 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 1-like [Jatropha curcas] |
| 13 |
Hb_147137_010 |
0.2514921324 |
- |
- |
hypothetical protein JCGZ_07229 [Jatropha curcas] |
| 14 |
Hb_000115_200 |
0.2538124885 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 15 |
Hb_000011_250 |
0.2545359037 |
- |
- |
PREDICTED: RING-H2 finger protein ATL3-like [Jatropha curcas] |
| 16 |
Hb_001157_210 |
0.2564053597 |
- |
- |
hypothetical protein CISIN_1g038260mg [Citrus sinensis] |
| 17 |
Hb_080192_010 |
0.2597774985 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 18 |
Hb_001102_310 |
0.2601949859 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 19 |
Hb_017914_020 |
0.2616553082 |
- |
- |
- |
| 20 |
Hb_007574_040 |
0.2626357619 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |