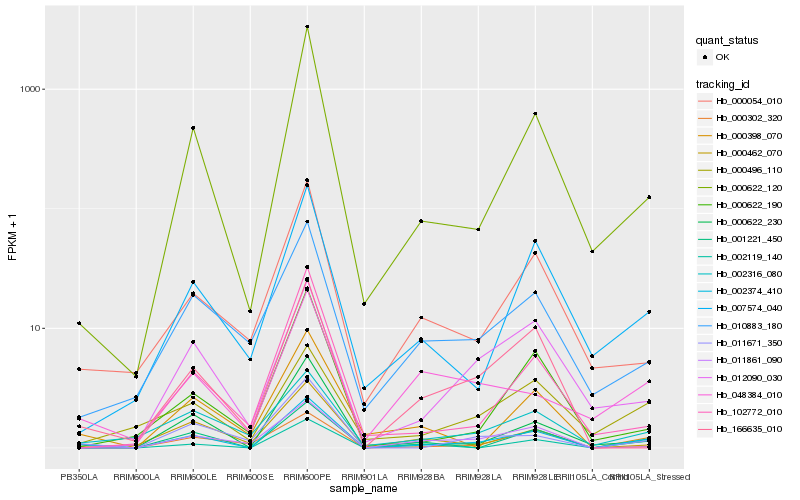
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_410 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_001221_450 |
0.17316804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000622_120 |
0.1770224304 |
- |
- |
hypothetical protein [Hevea brasiliensis] |
| 4 |
Hb_000622_230 |
0.2053105776 |
- |
- |
PREDICTED: uncharacterized protein LOC105642895 [Jatropha curcas] |
| 5 |
Hb_011671_350 |
0.2137010435 |
- |
- |
PREDICTED: uncharacterized protein LOC105648659 [Jatropha curcas] |
| 6 |
Hb_007574_040 |
0.2210859466 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 7 |
Hb_000054_010 |
0.223679857 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002316_080 |
0.2265161116 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 9 |
Hb_166635_010 |
0.2300142163 |
- |
- |
ptm1, putative [Ricinus communis] |
| 10 |
Hb_000462_070 |
0.2304555786 |
- |
- |
PREDICTED: probable pectinesterase 15 [Jatropha curcas] |
| 11 |
Hb_002119_140 |
0.2331037426 |
- |
- |
PREDICTED: uncharacterized protein LOC105634172 [Jatropha curcas] |
| 12 |
Hb_000622_190 |
0.2351193913 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |
| 13 |
Hb_012090_030 |
0.2351979085 |
- |
- |
PREDICTED: autophagy-related protein 8C isoform X2 [Phoenix dactylifera] |
| 14 |
Hb_010883_180 |
0.2369398103 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 15 |
Hb_102772_010 |
0.238571753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000398_070 |
0.2393463349 |
- |
- |
hypothetical protein VITISV_011880 [Vitis vinifera] |
| 17 |
Hb_048384_010 |
0.2398497968 |
- |
- |
PREDICTED: probable receptor-like protein kinase At2g42960 [Cucumis sativus] |
| 18 |
Hb_000302_320 |
0.2425558267 |
- |
- |
hypothetical protein JCGZ_26057 [Jatropha curcas] |
| 19 |
Hb_000496_110 |
0.2432290348 |
- |
- |
PREDICTED: uncharacterized protein LOC105650214 [Jatropha curcas] |
| 20 |
Hb_011861_090 |
0.2435390832 |
- |
- |
PREDICTED: probable sodium-coupled neutral amino acid transporter 6 [Jatropha curcas] |