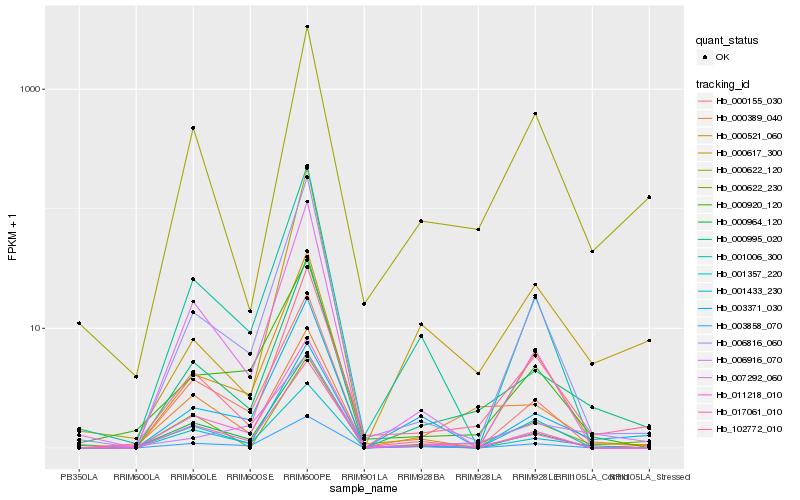
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000995_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_102772_010 |
0.1206433559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000622_230 |
0.1492035977 |
- |
- |
PREDICTED: uncharacterized protein LOC105642895 [Jatropha curcas] |
| 4 |
Hb_000622_120 |
0.1557921591 |
- |
- |
hypothetical protein [Hevea brasiliensis] |
| 5 |
Hb_006816_060 |
0.1575646391 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 6 |
Hb_003371_030 |
0.1580315449 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000617_300 |
0.162865827 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 8 |
Hb_001357_220 |
0.1667053895 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 9 |
Hb_006916_070 |
0.1687063316 |
transcription factor |
TF Family: MIKC |
PREDICTED: developmental protein SEPALLATA 1-like [Jatropha curcas] |
| 10 |
Hb_000521_060 |
0.1704026393 |
- |
- |
PREDICTED: protein YLS3-like [Jatropha curcas] |
| 11 |
Hb_011218_010 |
0.1716112256 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 12 |
Hb_000964_120 |
0.1722840361 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 13 |
Hb_000389_040 |
0.1739770442 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g03250-like [Jatropha curcas] |
| 14 |
Hb_000155_030 |
0.1756283497 |
- |
- |
PREDICTED: uncharacterized protein LOC105649591 [Jatropha curcas] |
| 15 |
Hb_017061_010 |
0.1764514884 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 16 |
Hb_001006_300 |
0.1770144958 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 17 |
Hb_000920_120 |
0.1783943826 |
- |
- |
- |
| 18 |
Hb_003858_070 |
0.1797326906 |
- |
- |
PREDICTED: uncharacterized protein LOC105647879 [Jatropha curcas] |
| 19 |
Hb_001433_230 |
0.1802958058 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 20 |
Hb_007292_060 |
0.1805605498 |
- |
- |
PREDICTED: uncharacterized protein LOC105632853 [Jatropha curcas] |