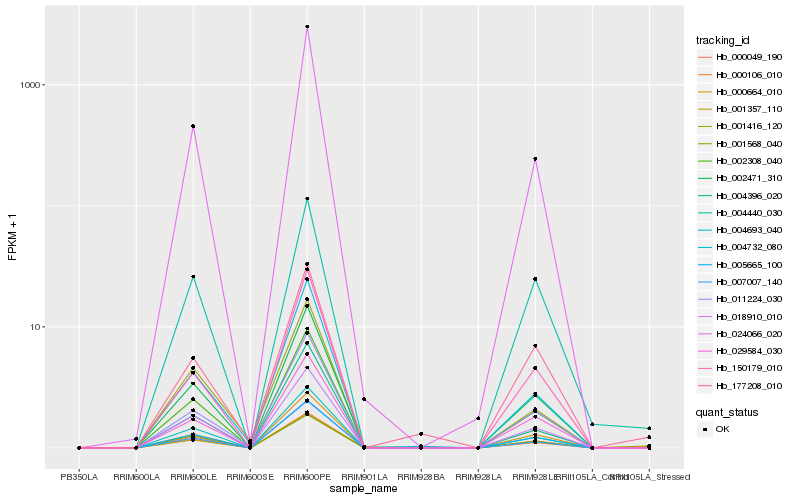
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000106_010 |
0.0 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 2 |
Hb_004693_040 |
0.10328722 |
- |
- |
- |
| 3 |
Hb_002308_040 |
0.103770462 |
- |
- |
methyladenine glycosylase family protein [Populus trichocarpa] |
| 4 |
Hb_002471_310 |
0.1063795911 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_018910_010 |
0.108242031 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180-like isoform X3 [Citrus sinensis] |
| 6 |
Hb_005665_100 |
0.1083350771 |
- |
- |
PREDICTED: RING-H2 finger protein ATL63-like [Populus euphratica] |
| 7 |
Hb_024066_020 |
0.1113621513 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 8 |
Hb_007007_140 |
0.1116040984 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL6 [Jatropha curcas] |
| 9 |
Hb_000664_010 |
0.1127141423 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 4-like [Jatropha curcas] |
| 10 |
Hb_000049_190 |
0.1173391905 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 11 |
Hb_001416_120 |
0.1178535913 |
- |
- |
caspase, putative [Ricinus communis] |
| 12 |
Hb_011224_030 |
0.1187135194 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_029584_030 |
0.1188980012 |
- |
- |
hypothetical protein JCGZ_07310 [Jatropha curcas] |
| 14 |
Hb_004440_030 |
0.1193918717 |
transcription factor |
TF Family: MYB-related |
hypothetical protein POPTR_0005s12390g [Populus trichocarpa] |
| 15 |
Hb_177208_010 |
0.1215402047 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor, partial [Jatropha sp. CONN 198700232] |
| 16 |
Hb_004396_020 |
0.1227884414 |
- |
- |
SAUR-like auxin-responsive protein family [Theobroma cacao] |
| 17 |
Hb_150179_010 |
0.1233232809 |
- |
- |
hypothetical protein JCGZ_21079 [Jatropha curcas] |
| 18 |
Hb_004732_080 |
0.1241396078 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 19 |
Hb_001357_110 |
0.1252958449 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 20 |
Hb_001568_040 |
0.1260058008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |