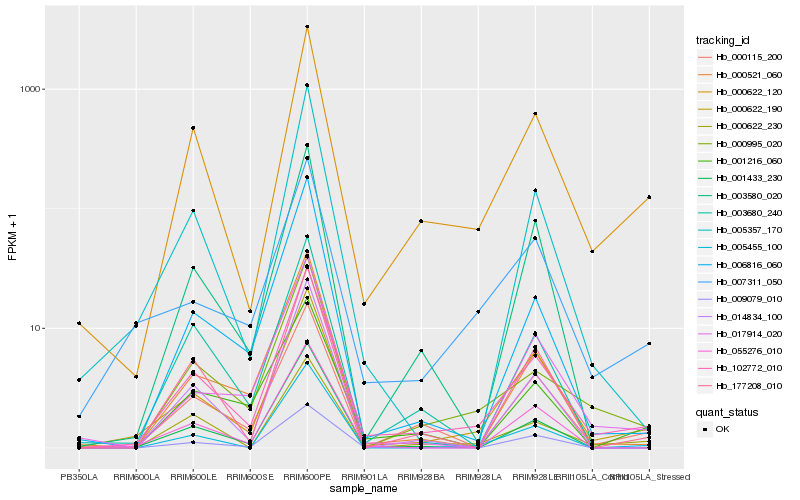
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_102772_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000622_120 |
0.0936932064 |
- |
- |
hypothetical protein [Hevea brasiliensis] |
| 3 |
Hb_000622_190 |
0.1038949807 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |
| 4 |
Hb_000995_020 |
0.1206433559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000622_230 |
0.1310933837 |
- |
- |
PREDICTED: uncharacterized protein LOC105642895 [Jatropha curcas] |
| 6 |
Hb_000115_200 |
0.1315627018 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 7 |
Hb_001433_230 |
0.1336264306 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 8 |
Hb_177208_010 |
0.1384150175 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor, partial [Jatropha sp. CONN 198700232] |
| 9 |
Hb_006816_060 |
0.1420912205 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 10 |
Hb_005455_100 |
0.143295232 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_000521_060 |
0.1437522362 |
- |
- |
PREDICTED: protein YLS3-like [Jatropha curcas] |
| 12 |
Hb_005357_170 |
0.145653479 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 13 |
Hb_055276_010 |
0.1467699359 |
- |
- |
hypothetical protein POPTR_0003s02950g [Populus trichocarpa] |
| 14 |
Hb_007311_050 |
0.1484787648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003580_020 |
0.1489490921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001216_060 |
0.1489535415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_014834_100 |
0.1493336302 |
- |
- |
PREDICTED: probable aquaporin TIP-type [Gossypium raimondii] |
| 18 |
Hb_003680_240 |
0.1506350836 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 19 |
Hb_017914_020 |
0.1525412694 |
- |
- |
- |
| 20 |
Hb_009079_010 |
0.1540670444 |
- |
- |
JHL18I08.8 [Jatropha curcas] |