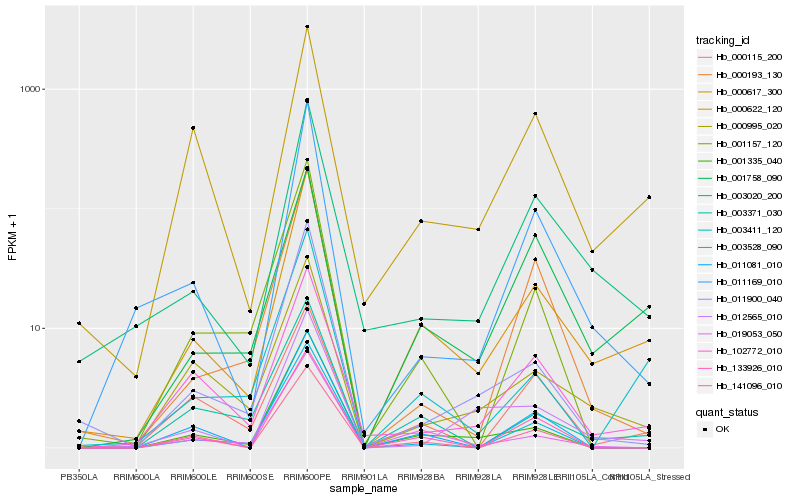
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000617_300 |
0.0 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 2 |
Hb_003020_200 |
0.1423916026 |
- |
- |
PREDICTED: uncharacterized protein LOC105131117 [Populus euphratica] |
| 3 |
Hb_003411_120 |
0.1520962044 |
- |
- |
hypothetical protein POPTR_0010s13670g [Populus trichocarpa] |
| 4 |
Hb_001758_090 |
0.1527298834 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 5 |
Hb_003371_030 |
0.157561798 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 6 |
Hb_019053_050 |
0.1584038743 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_000995_020 |
0.162865827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001335_040 |
0.1629747653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_102772_010 |
0.166282078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_011169_010 |
0.1669644882 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 11 |
Hb_000622_120 |
0.169131359 |
- |
- |
hypothetical protein [Hevea brasiliensis] |
| 12 |
Hb_011900_040 |
0.1746382621 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 13 |
Hb_000115_200 |
0.1796413688 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 14 |
Hb_133926_010 |
0.1796927133 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 15 |
Hb_011081_010 |
0.1826926433 |
- |
- |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_003528_090 |
0.1843230392 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |
| 17 |
Hb_141096_010 |
0.1861425005 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 55-like [Jatropha curcas] |
| 18 |
Hb_012565_010 |
0.1884554718 |
- |
- |
PREDICTED: putative glutamine amidotransferase PB2B2.05 [Jatropha curcas] |
| 19 |
Hb_001157_120 |
0.1889055149 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 20 |
Hb_000193_130 |
0.1889688847 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |