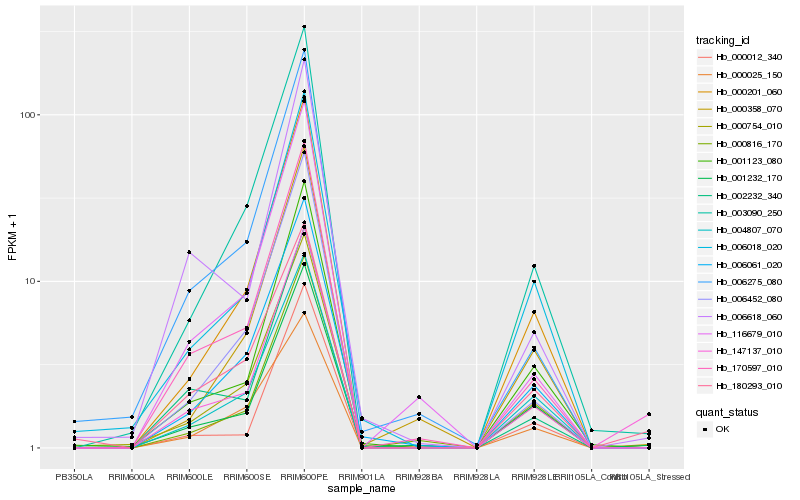
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_147137_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_07229 [Jatropha curcas] |
| 2 |
Hb_180293_010 |
0.1079410779 |
- |
- |
hypothetical protein POPTR_0008s19020g [Populus trichocarpa] |
| 3 |
Hb_001232_170 |
0.1158757082 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 4 |
Hb_170597_010 |
0.1227341553 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 5 |
Hb_003090_250 |
0.1253885086 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_000201_060 |
0.1272678202 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X2 [Populus euphratica] |
| 7 |
Hb_000025_150 |
0.1304141964 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 8 |
Hb_002232_340 |
0.1311757194 |
- |
- |
Sodium transporter hkt1-like protein, putative [Theobroma cacao] |
| 9 |
Hb_000754_010 |
0.1316960342 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_001123_080 |
0.1320074662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004807_070 |
0.1349549957 |
- |
- |
hypothetical protein POPTR_0004s17330g, partial [Populus trichocarpa] |
| 12 |
Hb_006452_080 |
0.1363374779 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 13 |
Hb_006061_020 |
0.1371147758 |
transcription factor |
TF Family: bZIP |
bZIP family protein [Populus trichocarpa] |
| 14 |
Hb_006018_020 |
0.1388141115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000816_170 |
0.1399304618 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 15 [Jatropha curcas] |
| 16 |
Hb_116679_010 |
0.141880308 |
- |
- |
PREDICTED: transcription factor bHLH110 isoform X2 [Vitis vinifera] |
| 17 |
Hb_000012_340 |
0.1425943545 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |
| 18 |
Hb_000358_070 |
0.1427271939 |
- |
- |
hypothetical protein POPTR_0003s01410g [Populus trichocarpa] |
| 19 |
Hb_006618_060 |
0.1437837624 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 20 |
Hb_006275_080 |
0.1443099997 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |