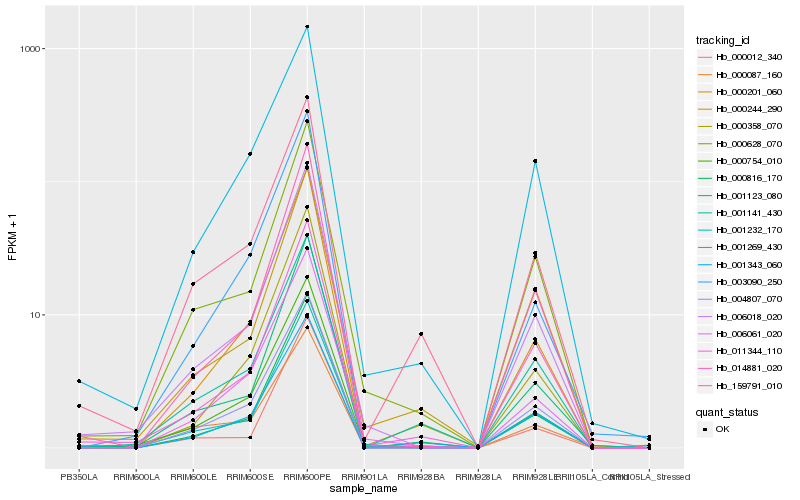
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006018_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001123_080 |
0.0492577457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000754_010 |
0.050058932 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_001232_170 |
0.0600635721 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 5 |
Hb_014881_020 |
0.0622949798 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 6 |
Hb_000628_070 |
0.0645532316 |
- |
- |
- |
| 7 |
Hb_011344_110 |
0.0707833377 |
transcription factor |
TF Family: MYB |
PREDICTED: protein ODORANT1 [Jatropha curcas] |
| 8 |
Hb_004807_070 |
0.0741674292 |
- |
- |
hypothetical protein POPTR_0004s17330g, partial [Populus trichocarpa] |
| 9 |
Hb_000201_060 |
0.0743187973 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X2 [Populus euphratica] |
| 10 |
Hb_006061_020 |
0.0774274643 |
transcription factor |
TF Family: bZIP |
bZIP family protein [Populus trichocarpa] |
| 11 |
Hb_000816_170 |
0.0848477437 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 15 [Jatropha curcas] |
| 12 |
Hb_000244_290 |
0.0892125907 |
- |
- |
PREDICTED: UDP-glucuronate:xylan alpha-glucuronosyltransferase 1 [Jatropha curcas] |
| 13 |
Hb_001269_430 |
0.0919215487 |
- |
- |
hypothetical protein JCGZ_04286 [Jatropha curcas] |
| 14 |
Hb_003090_250 |
0.0925654813 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_001343_060 |
0.0984237122 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 16 |
Hb_000358_070 |
0.0989180078 |
- |
- |
hypothetical protein POPTR_0003s01410g [Populus trichocarpa] |
| 17 |
Hb_159791_010 |
0.099200814 |
- |
- |
PREDICTED: cytochrome P450 84A1 [Jatropha curcas] |
| 18 |
Hb_000012_340 |
0.1000909772 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |
| 19 |
Hb_000087_160 |
0.1019817177 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_001141_430 |
0.1027895759 |
- |
- |
hypothetical protein POPTR_0008s15160g [Populus trichocarpa] |