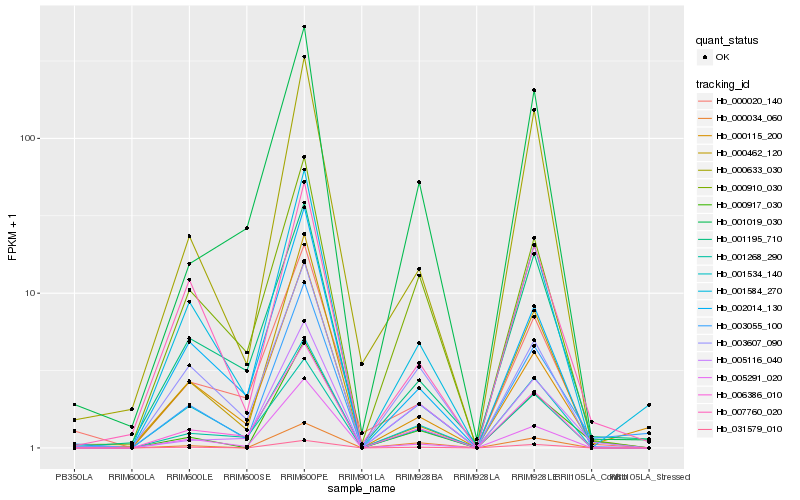
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003055_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 2 |
Hb_001584_270 |
0.1015868761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000115_200 |
0.1223539765 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 4 |
Hb_000917_030 |
0.1307671797 |
- |
- |
PREDICTED: uncharacterized protein LOC102629571 [Citrus sinensis] |
| 5 |
Hb_006386_010 |
0.1384078195 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 6 |
Hb_001195_710 |
0.1424962813 |
- |
- |
glycosyltransferase family GT8 protein [Populus deltoides] |
| 7 |
Hb_031579_010 |
0.1457652155 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 8 |
Hb_000910_030 |
0.1502312353 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007760_020 |
0.1534672303 |
- |
- |
PREDICTED: expansin-A1 [Populus euphratica] |
| 10 |
Hb_001268_290 |
0.1538083736 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 11 |
Hb_000020_140 |
0.154368069 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 12 |
Hb_005291_020 |
0.1545510093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000633_030 |
0.1561849954 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 14 |
Hb_001019_030 |
0.157215205 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 15 |
Hb_000034_060 |
0.1576805863 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Citrus sinensis] |
| 16 |
Hb_003607_090 |
0.1594225706 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 17 |
Hb_005116_040 |
0.1600024142 |
- |
- |
PREDICTED: uncharacterized protein LOC105131445 [Populus euphratica] |
| 18 |
Hb_002014_130 |
0.1608728825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000462_120 |
0.1616541645 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 20 |
Hb_001534_140 |
0.1652729065 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |