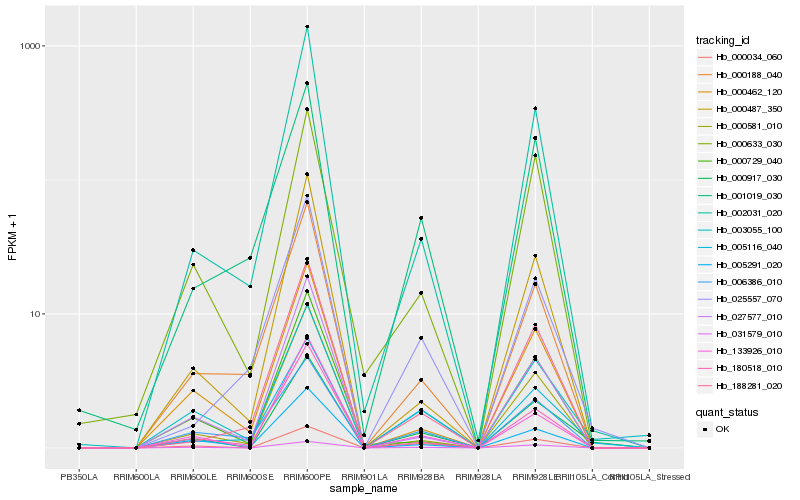
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000917_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102629571 [Citrus sinensis] |
| 2 |
Hb_031579_010 |
0.1273737068 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 3 |
Hb_003055_100 |
0.1307671797 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 4 |
Hb_000034_060 |
0.1370454759 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Citrus sinensis] |
| 5 |
Hb_005116_040 |
0.1386215987 |
- |
- |
PREDICTED: uncharacterized protein LOC105131445 [Populus euphratica] |
| 6 |
Hb_002031_020 |
0.1496431545 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 7 |
Hb_000633_030 |
0.1517179687 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 8 |
Hb_005291_020 |
0.1572724507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000462_120 |
0.161590962 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 10 |
Hb_000581_010 |
0.1625080662 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 11 |
Hb_133926_010 |
0.1632085731 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 12 |
Hb_180518_010 |
0.1643520809 |
- |
- |
PREDICTED: receptor-like protein 2 [Jatropha curcas] |
| 13 |
Hb_000487_350 |
0.1649304765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001019_030 |
0.1658105869 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 15 |
Hb_006386_010 |
0.1663766287 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 16 |
Hb_025557_070 |
0.1686215156 |
transcription factor |
TF Family: zf-HD |
hypothetical protein JCGZ_11626 [Jatropha curcas] |
| 17 |
Hb_188281_020 |
0.1696084461 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 18 |
Hb_000729_040 |
0.1697797615 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 10 precursor [Populus trichocarpa] |
| 19 |
Hb_027577_010 |
0.1703576404 |
- |
- |
PREDICTED: uncharacterized protein LOC104779928 [Camelina sativa] |
| 20 |
Hb_000188_040 |
0.171183043 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |