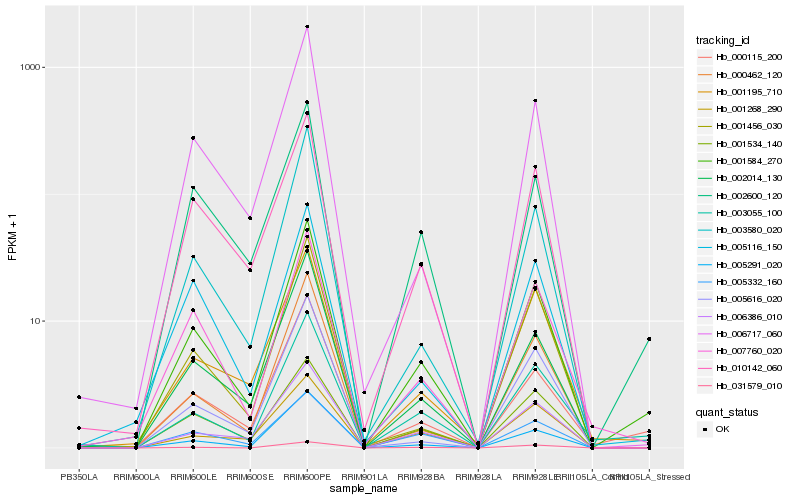
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001584_270 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000115_200 |
0.0943649125 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 3 |
Hb_003055_100 |
0.1015868761 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 4 |
Hb_002600_120 |
0.105997295 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 5 |
Hb_002014_130 |
0.1071605429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_006386_010 |
0.1094679287 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 7 |
Hb_005291_020 |
0.1098899524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001534_140 |
0.1123382361 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 9 |
Hb_007760_020 |
0.1152480979 |
- |
- |
PREDICTED: expansin-A1 [Populus euphratica] |
| 10 |
Hb_010142_060 |
0.1158767214 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 11 |
Hb_000462_120 |
0.118028129 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 12 |
Hb_001195_710 |
0.119072532 |
- |
- |
glycosyltransferase family GT8 protein [Populus deltoides] |
| 13 |
Hb_003580_020 |
0.1197914831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006717_060 |
0.1226668453 |
- |
- |
PREDICTED: peroxidase 42 [Jatropha curcas] |
| 15 |
Hb_005116_150 |
0.1264535969 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005332_160 |
0.1306914701 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 2-like [Jatropha curcas] |
| 17 |
Hb_031579_010 |
0.1308893469 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 18 |
Hb_001456_030 |
0.131789846 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |
| 19 |
Hb_001268_290 |
0.1348486228 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 20 |
Hb_005616_020 |
0.1357178803 |
- |
- |
kinase, putative [Ricinus communis] |