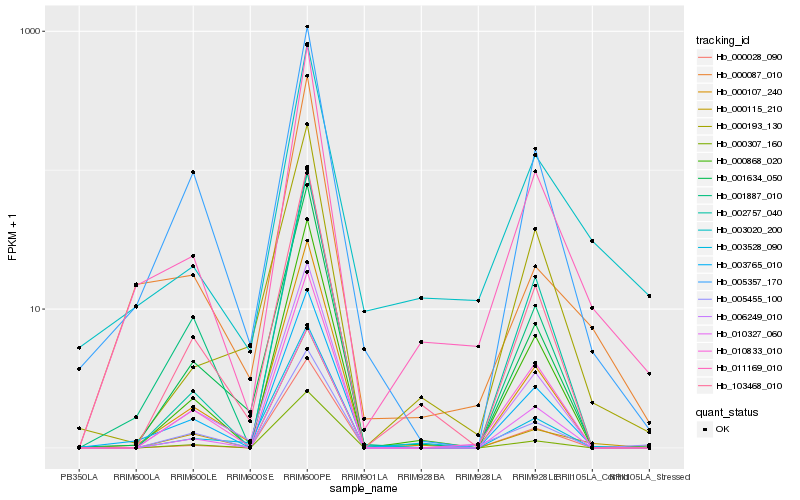
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011169_010 |
0.0 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 2 |
Hb_003765_010 |
0.1058613921 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 3 |
Hb_000107_240 |
0.1208023249 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 4 |
Hb_003020_200 |
0.1231337867 |
- |
- |
PREDICTED: uncharacterized protein LOC105131117 [Populus euphratica] |
| 5 |
Hb_000868_020 |
0.1294961749 |
- |
- |
- |
| 6 |
Hb_000087_010 |
0.129663196 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 7 |
Hb_103468_010 |
0.1381394591 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_005357_170 |
0.1381419593 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 9 |
Hb_000193_130 |
0.1418357617 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_001634_050 |
0.1421396133 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 11 |
Hb_010327_060 |
0.1421903923 |
- |
- |
hypothetical protein B456_001G246300 [Gossypium raimondii] |
| 12 |
Hb_010833_010 |
0.142680671 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |
| 13 |
Hb_006249_010 |
0.1439037765 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 14 |
Hb_000028_090 |
0.1453498842 |
- |
- |
PREDICTED: uncharacterized protein LOC105135810 [Populus euphratica] |
| 15 |
Hb_001887_010 |
0.1456330023 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 16 |
Hb_000115_210 |
0.1476056351 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 3-like [Jatropha curcas] |
| 17 |
Hb_002757_040 |
0.1477161166 |
- |
- |
- |
| 18 |
Hb_003528_090 |
0.148037188 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |
| 19 |
Hb_000307_160 |
0.1480604543 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 20 |
Hb_005455_100 |
0.1488661063 |
- |
- |
amino acid transporter, putative [Ricinus communis] |