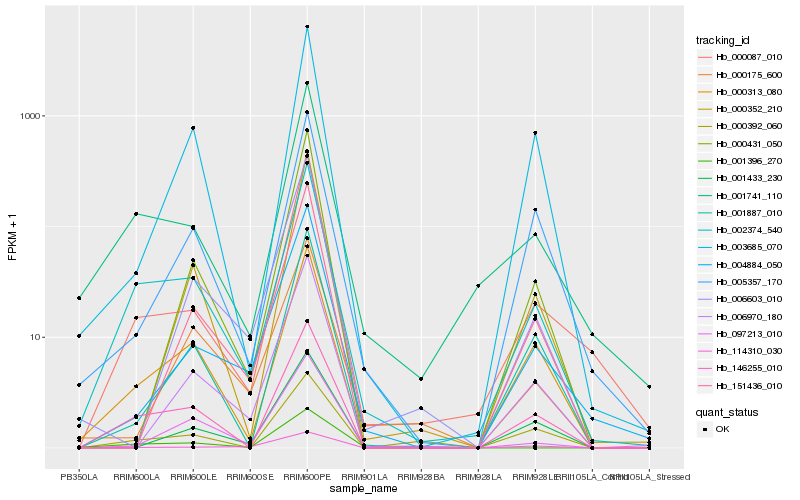
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_540 |
0.0 |
- |
- |
RecName: Full=Esterase; AltName: Full=Early nodule-specific protein homolog; AltName: Full=Latex allergen Hev b 13; AltName: Allergen=Hev b 13; Flags: Precursor [Hevea brasiliensis] |
| 2 |
Hb_146255_010 |
0.0881376922 |
- |
- |
PREDICTED: uncharacterized protein LOC105643649 [Jatropha curcas] |
| 3 |
Hb_001741_110 |
0.1128567039 |
rubber biosynthesis |
Gene Name: REF7 |
REF/SRPP-like protein [Glycine soja] |
| 4 |
Hb_000313_080 |
0.1273129503 |
rubber biosynthesis |
Gene Name: CPT3 |
cis-prenyltransferase 3 [Hevea brasiliensis] |
| 5 |
Hb_000087_010 |
0.1417764372 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 6 |
Hb_001396_270 |
0.174989156 |
- |
- |
4-coumarate--CoA ligase family protein [Populus trichocarpa] |
| 7 |
Hb_005357_170 |
0.1758171923 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 8 |
Hb_001887_010 |
0.1765274781 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 9 |
Hb_004884_050 |
0.1769211219 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 10 |
Hb_003685_070 |
0.180863896 |
- |
- |
RecName: Full=(S)-hydroxynitrile lyase; AltName: Full=(S)-acetone-cyanohydrin lyase; AltName: Full=Oxynitrilase [Hevea brasiliensis] |
| 11 |
Hb_000392_060 |
0.1809385235 |
- |
- |
hypothetical protein JCGZ_11629 [Jatropha curcas] |
| 12 |
Hb_097213_010 |
0.1830887658 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Populus euphratica] |
| 13 |
Hb_006970_180 |
0.1884121678 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 14 |
Hb_114310_030 |
0.1893989705 |
- |
- |
JHL18I08.8 [Jatropha curcas] |
| 15 |
Hb_151436_010 |
0.1899411447 |
- |
- |
PREDICTED: uncharacterized protein LOC105137019 [Populus euphratica] |
| 16 |
Hb_001433_230 |
0.1900095204 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 17 |
Hb_000352_210 |
0.1917911613 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 18 |
Hb_000431_050 |
0.1918818261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_006603_010 |
0.1921631609 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 20 |
Hb_000175_600 |
0.19220172 |
- |
- |
PREDICTED: TATA-binding protein-associated factor 2N isoform X2 [Jatropha curcas] |