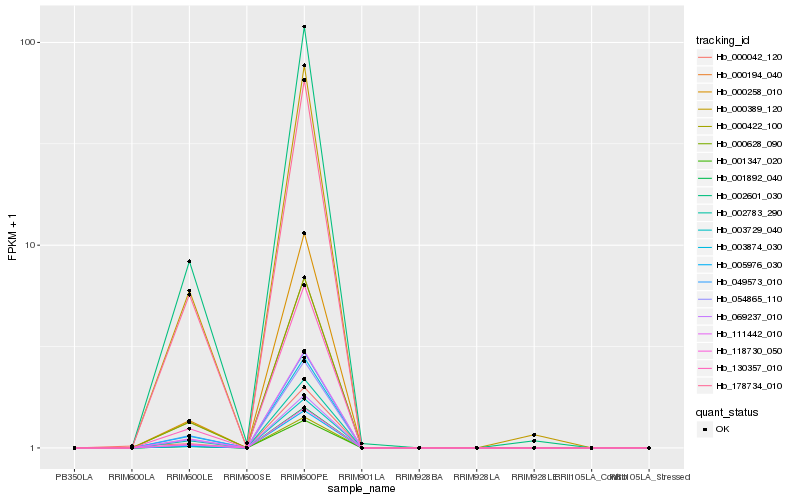
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632800 [Jatropha curcas] |
| 2 |
Hb_001347_020 |
0.1012061056 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Populus euphratica] |
| 3 |
Hb_000422_100 |
0.1012513187 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 4 |
Hb_111442_010 |
0.1014092319 |
desease resistance |
Gene Name: AAA_16 |
LRR and NB-ARC domains-containing disease resistance protein, putative [Theobroma cacao] |
| 5 |
Hb_130357_010 |
0.1014447047 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 6 |
Hb_003729_040 |
0.1018442604 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000628_090 |
0.1019367523 |
- |
- |
hypothetical protein B456_010G019400 [Gossypium raimondii] |
| 8 |
Hb_054865_110 |
0.1020990075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000194_040 |
0.1021629377 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 10 |
Hb_069237_010 |
0.1026819493 |
- |
- |
- |
| 11 |
Hb_000258_010 |
0.1054715846 |
- |
- |
lipid transfer precursor protein [Hevea brasiliensis] |
| 12 |
Hb_002601_030 |
0.1067550239 |
- |
- |
PREDICTED: uncharacterized protein LOC105634908 [Jatropha curcas] |
| 13 |
Hb_118730_050 |
0.1068964419 |
- |
- |
RING/U-box superfamily protein isoform 4 [Theobroma cacao] |
| 14 |
Hb_002783_290 |
0.1069382482 |
- |
- |
Uncharacterized protein TCM_034216 [Theobroma cacao] |
| 15 |
Hb_005976_030 |
0.1069969695 |
- |
- |
PREDICTED: uncharacterized protein LOC105639715 [Jatropha curcas] |
| 16 |
Hb_178734_010 |
0.1079313974 |
- |
- |
PREDICTED: uncharacterized protein LOC103331175 [Prunus mume] |
| 17 |
Hb_000389_120 |
0.1089773405 |
- |
- |
hypothetical protein JCGZ_20066 [Jatropha curcas] |
| 18 |
Hb_049573_010 |
0.1093737633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003874_030 |
0.1113953037 |
- |
- |
hypothetical protein JCGZ_00004 [Jatropha curcas] |
| 20 |
Hb_001892_040 |
0.1117889812 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |