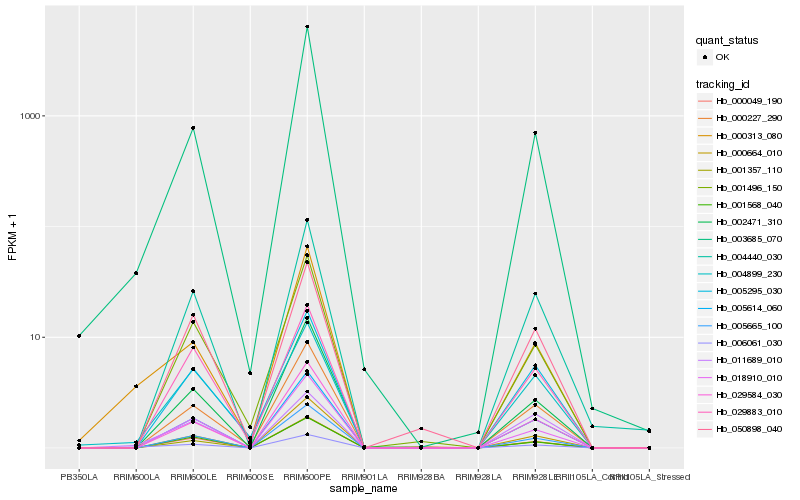
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006061_030 |
0.0 |
- |
- |
PREDICTED: fe(2+) transport protein 1-like [Jatropha curcas] |
| 2 |
Hb_000313_080 |
0.1174908535 |
rubber biosynthesis |
Gene Name: CPT3 |
cis-prenyltransferase 3 [Hevea brasiliensis] |
| 3 |
Hb_005614_060 |
0.1357277573 |
- |
- |
Uncharacterized protein TCM_019750 [Theobroma cacao] |
| 4 |
Hb_000049_190 |
0.1389788637 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 5 |
Hb_005665_100 |
0.1415001367 |
- |
- |
PREDICTED: RING-H2 finger protein ATL63-like [Populus euphratica] |
| 6 |
Hb_004899_230 |
0.1415731241 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 7 |
Hb_001568_040 |
0.1437910727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_018910_010 |
0.1461376346 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180-like isoform X3 [Citrus sinensis] |
| 9 |
Hb_004440_030 |
0.1464699345 |
transcription factor |
TF Family: MYB-related |
hypothetical protein POPTR_0005s12390g [Populus trichocarpa] |
| 10 |
Hb_011689_010 |
0.1500245578 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 11 |
Hb_000664_010 |
0.1510251778 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 4-like [Jatropha curcas] |
| 12 |
Hb_000227_290 |
0.1519986304 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 13 |
Hb_029584_030 |
0.1550265233 |
- |
- |
hypothetical protein JCGZ_07310 [Jatropha curcas] |
| 14 |
Hb_001357_110 |
0.1554373278 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 15 |
Hb_050898_040 |
0.1560873906 |
- |
- |
- |
| 16 |
Hb_001496_150 |
0.1570300507 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 17 |
Hb_002471_310 |
0.157330923 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_029883_010 |
0.1575738991 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 19 |
Hb_003685_070 |
0.1578348894 |
- |
- |
RecName: Full=(S)-hydroxynitrile lyase; AltName: Full=(S)-acetone-cyanohydrin lyase; AltName: Full=Oxynitrilase [Hevea brasiliensis] |
| 20 |
Hb_005295_030 |
0.158414008 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |