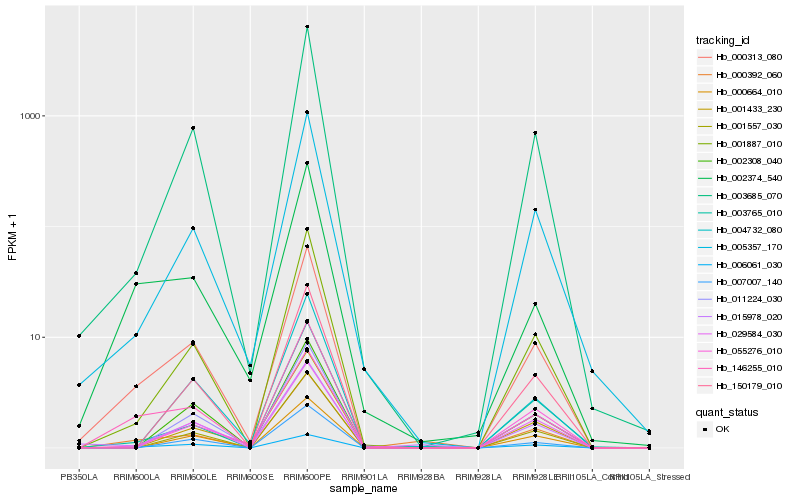
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000313_080 |
0.0 |
rubber biosynthesis |
Gene Name: CPT3 |
cis-prenyltransferase 3 [Hevea brasiliensis] |
| 2 |
Hb_146255_010 |
0.0867468367 |
- |
- |
PREDICTED: uncharacterized protein LOC105643649 [Jatropha curcas] |
| 3 |
Hb_003685_070 |
0.0940746639 |
- |
- |
RecName: Full=(S)-hydroxynitrile lyase; AltName: Full=(S)-acetone-cyanohydrin lyase; AltName: Full=Oxynitrilase [Hevea brasiliensis] |
| 4 |
Hb_005357_170 |
0.104829101 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 5 |
Hb_001887_010 |
0.1073348824 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 6 |
Hb_006061_030 |
0.1174908535 |
- |
- |
PREDICTED: fe(2+) transport protein 1-like [Jatropha curcas] |
| 7 |
Hb_015978_020 |
0.1241187568 |
- |
- |
Multidrug resistance protein ABC transporter family protein, putative [Theobroma cacao] |
| 8 |
Hb_002374_540 |
0.1273129503 |
- |
- |
RecName: Full=Esterase; AltName: Full=Early nodule-specific protein homolog; AltName: Full=Latex allergen Hev b 13; AltName: Allergen=Hev b 13; Flags: Precursor [Hevea brasiliensis] |
| 9 |
Hb_011224_030 |
0.1297501799 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_150179_010 |
0.1331995224 |
- |
- |
hypothetical protein JCGZ_21079 [Jatropha curcas] |
| 11 |
Hb_055276_010 |
0.1355790515 |
- |
- |
hypothetical protein POPTR_0003s02950g [Populus trichocarpa] |
| 12 |
Hb_003765_010 |
0.1355857253 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 13 |
Hb_029584_030 |
0.1389606647 |
- |
- |
hypothetical protein JCGZ_07310 [Jatropha curcas] |
| 14 |
Hb_000664_010 |
0.1391401333 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 4-like [Jatropha curcas] |
| 15 |
Hb_007007_140 |
0.1404595145 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL6 [Jatropha curcas] |
| 16 |
Hb_001557_030 |
0.1406938596 |
- |
- |
hypothetical protein CICLE_v10005306mg [Citrus clementina] |
| 17 |
Hb_000392_060 |
0.1409560355 |
- |
- |
hypothetical protein JCGZ_11629 [Jatropha curcas] |
| 18 |
Hb_001433_230 |
0.141342696 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 19 |
Hb_004732_080 |
0.1421408755 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 20 |
Hb_002308_040 |
0.14268069 |
- |
- |
methyladenine glycosylase family protein [Populus trichocarpa] |