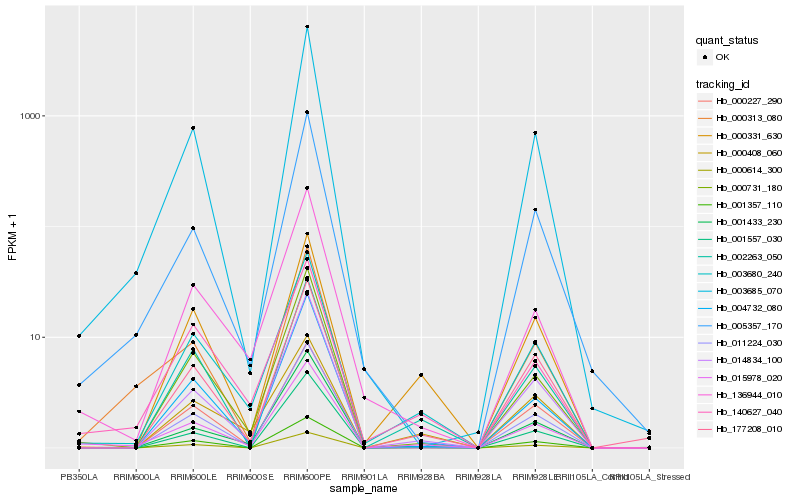
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015978_020 |
0.0 |
- |
- |
Multidrug resistance protein ABC transporter family protein, putative [Theobroma cacao] |
| 2 |
Hb_003680_240 |
0.0938686654 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 3 |
Hb_003685_070 |
0.1109934735 |
- |
- |
RecName: Full=(S)-hydroxynitrile lyase; AltName: Full=(S)-acetone-cyanohydrin lyase; AltName: Full=Oxynitrilase [Hevea brasiliensis] |
| 4 |
Hb_014834_100 |
0.1121338037 |
- |
- |
PREDICTED: probable aquaporin TIP-type [Gossypium raimondii] |
| 5 |
Hb_140627_040 |
0.115155062 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 6 |
Hb_001433_230 |
0.1152648871 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 7 |
Hb_005357_170 |
0.1195289604 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 8 |
Hb_000731_180 |
0.120114767 |
- |
- |
hypothetical protein POPTR_0004s01820g [Populus trichocarpa] |
| 9 |
Hb_001357_110 |
0.1229144374 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 10 |
Hb_000227_290 |
0.1236732342 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 11 |
Hb_000313_080 |
0.1241187568 |
rubber biosynthesis |
Gene Name: CPT3 |
cis-prenyltransferase 3 [Hevea brasiliensis] |
| 12 |
Hb_136944_010 |
0.1245175603 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 13 |
Hb_011224_030 |
0.1251568135 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_177208_010 |
0.1265595922 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor, partial [Jatropha sp. CONN 198700232] |
| 15 |
Hb_001557_030 |
0.1274909944 |
- |
- |
hypothetical protein CICLE_v10005306mg [Citrus clementina] |
| 16 |
Hb_000614_300 |
0.127814183 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Jatropha curcas] |
| 17 |
Hb_002263_050 |
0.1282774241 |
- |
- |
hypothetical protein POPTR_0016s06160g [Populus trichocarpa] |
| 18 |
Hb_004732_080 |
0.1285163281 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 19 |
Hb_000331_630 |
0.1304176911 |
- |
- |
hypothetical protein B456_008G203900 [Gossypium raimondii] |
| 20 |
Hb_000408_060 |
0.1313473349 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |