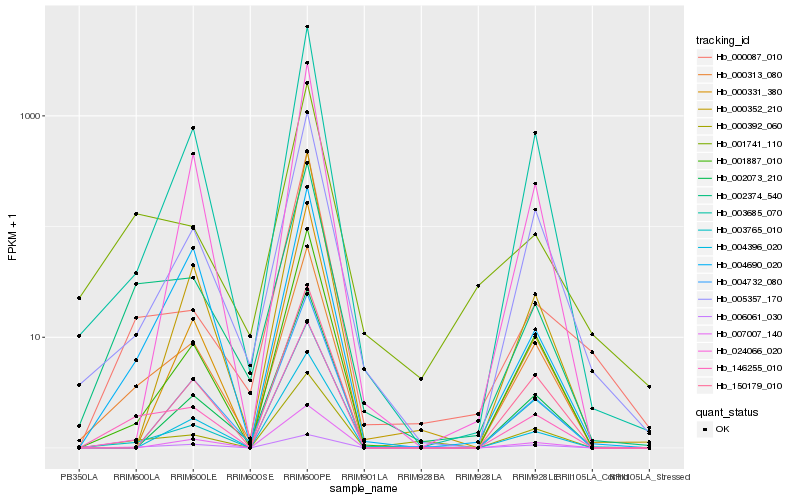
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146255_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643649 [Jatropha curcas] |
| 2 |
Hb_000313_080 |
0.0867468367 |
rubber biosynthesis |
Gene Name: CPT3 |
cis-prenyltransferase 3 [Hevea brasiliensis] |
| 3 |
Hb_002374_540 |
0.0881376922 |
- |
- |
RecName: Full=Esterase; AltName: Full=Early nodule-specific protein homolog; AltName: Full=Latex allergen Hev b 13; AltName: Allergen=Hev b 13; Flags: Precursor [Hevea brasiliensis] |
| 4 |
Hb_001887_010 |
0.1410877632 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 5 |
Hb_000392_060 |
0.1470501928 |
- |
- |
hypothetical protein JCGZ_11629 [Jatropha curcas] |
| 6 |
Hb_003685_070 |
0.1498471789 |
- |
- |
RecName: Full=(S)-hydroxynitrile lyase; AltName: Full=(S)-acetone-cyanohydrin lyase; AltName: Full=Oxynitrilase [Hevea brasiliensis] |
| 7 |
Hb_006061_030 |
0.1593683131 |
- |
- |
PREDICTED: fe(2+) transport protein 1-like [Jatropha curcas] |
| 8 |
Hb_001741_110 |
0.159923194 |
rubber biosynthesis |
Gene Name: REF7 |
REF/SRPP-like protein [Glycine soja] |
| 9 |
Hb_005357_170 |
0.1625288098 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 10 |
Hb_000087_010 |
0.1667467262 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 11 |
Hb_003765_010 |
0.1699116863 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 12 |
Hb_007007_140 |
0.1752510921 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL6 [Jatropha curcas] |
| 13 |
Hb_000331_380 |
0.1752670423 |
- |
- |
hypothetical protein POPTR_0012s09560g [Populus trichocarpa] |
| 14 |
Hb_004396_020 |
0.1755311065 |
- |
- |
SAUR-like auxin-responsive protein family [Theobroma cacao] |
| 15 |
Hb_024066_020 |
0.1777331847 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 16 |
Hb_150179_010 |
0.1790745269 |
- |
- |
hypothetical protein JCGZ_21079 [Jatropha curcas] |
| 17 |
Hb_000352_210 |
0.1792471714 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 18 |
Hb_002073_210 |
0.1798769687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004732_080 |
0.1799492199 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 20 |
Hb_004690_020 |
0.1802777416 |
- |
- |
PREDICTED: MLP-like protein 328 [Jatropha curcas] |