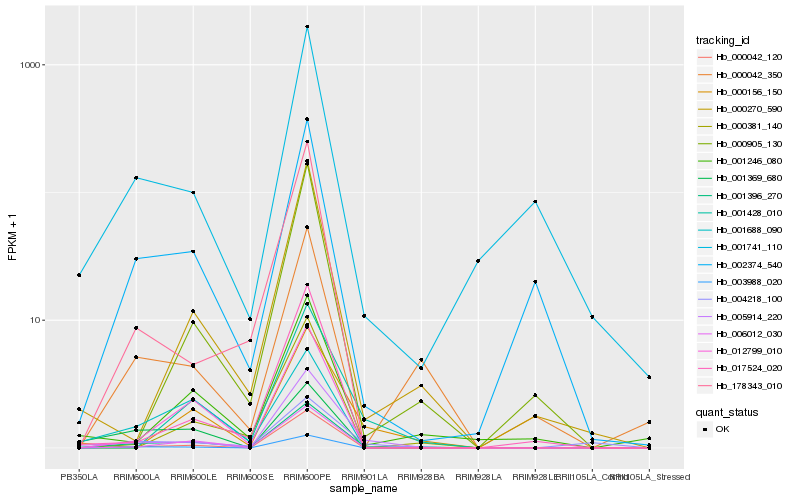
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001396_270 |
0.0 |
- |
- |
4-coumarate--CoA ligase family protein [Populus trichocarpa] |
| 2 |
Hb_006012_030 |
0.1251977084 |
- |
- |
hypothetical protein POPTR_0008s02380g [Populus trichocarpa] |
| 3 |
Hb_005914_220 |
0.145017699 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003988_020 |
0.1597239456 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH85-like [Populus euphratica] |
| 5 |
Hb_000042_120 |
0.1714291497 |
- |
- |
PREDICTED: uncharacterized protein LOC105632800 [Jatropha curcas] |
| 6 |
Hb_002374_540 |
0.174989156 |
- |
- |
RecName: Full=Esterase; AltName: Full=Early nodule-specific protein homolog; AltName: Full=Latex allergen Hev b 13; AltName: Allergen=Hev b 13; Flags: Precursor [Hevea brasiliensis] |
| 7 |
Hb_000270_590 |
0.1807553673 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 8 |
Hb_012799_010 |
0.1822996047 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001741_110 |
0.1860192259 |
rubber biosynthesis |
Gene Name: REF7 |
REF/SRPP-like protein [Glycine soja] |
| 10 |
Hb_000042_350 |
0.1906678504 |
- |
- |
hypothetical protein JCGZ_00710 [Jatropha curcas] |
| 11 |
Hb_001246_080 |
0.1941141168 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 12 |
Hb_000156_150 |
0.1951878318 |
- |
- |
hypothetical protein JCGZ_04603 [Jatropha curcas] |
| 13 |
Hb_001428_010 |
0.1969585301 |
- |
- |
PREDICTED: transcription factor PRE1-like [Jatropha curcas] |
| 14 |
Hb_017524_020 |
0.1995729384 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 15 |
Hb_178343_010 |
0.2001642947 |
- |
- |
PREDICTED: two-component response regulator ARR17-like [Jatropha curcas] |
| 16 |
Hb_001688_090 |
0.2021999777 |
- |
- |
alcohol dehydrogenase 7 [Vitis vinifera] |
| 17 |
Hb_001369_680 |
0.2038133229 |
- |
- |
PREDICTED: uncharacterized protein LOC105648582 [Jatropha curcas] |
| 18 |
Hb_000905_130 |
0.205585454 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100 [Jatropha curcas] |
| 19 |
Hb_000381_140 |
0.2061909223 |
- |
- |
PREDICTED: S-type anion channel SLAH4-like [Jatropha curcas] |
| 20 |
Hb_004218_100 |
0.2078156293 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 5C [Jatropha curcas] |