| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
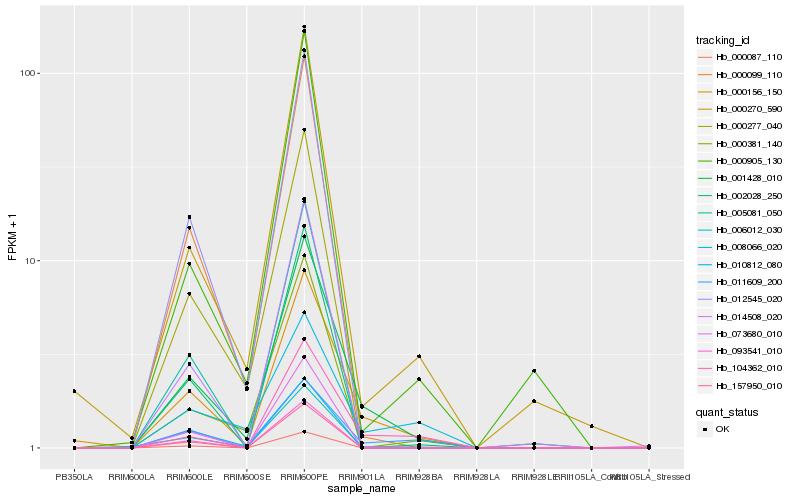
Hb_001428_010 |
0.0 |
- |
- |
PREDICTED: transcription factor PRE1-like [Jatropha curcas] |
| 2 |
Hb_000156_150 |
0.0821607546 |
- |
- |
hypothetical protein JCGZ_04603 [Jatropha curcas] |
| 3 |
Hb_006012_030 |
0.1275746644 |
- |
- |
hypothetical protein POPTR_0008s02380g [Populus trichocarpa] |
| 4 |
Hb_104362_010 |
0.1472202904 |
- |
- |
hypothetical protein POPTR_0247s002302g, partial [Populus trichocarpa] |
| 5 |
Hb_000099_110 |
0.1550485668 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 6 |
Hb_011609_200 |
0.160009466 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8-like isoform X3 [Populus euphratica] |
| 7 |
Hb_000270_590 |
0.1619087602 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 8 |
Hb_012545_020 |
0.1655391447 |
- |
- |
PREDICTED: uncharacterized protein LOC105633903 [Jatropha curcas] |
| 9 |
Hb_000277_040 |
0.1697574891 |
- |
- |
- |
| 10 |
Hb_000381_140 |
0.1717981142 |
- |
- |
PREDICTED: S-type anion channel SLAH4-like [Jatropha curcas] |
| 11 |
Hb_014508_020 |
0.1746530277 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002028_250 |
0.1749544334 |
- |
- |
Alpha-aminoadipic semialdehyde synthase, mitochondrial [Crassostrea gigas] |
| 13 |
Hb_000905_130 |
0.1761127724 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100 [Jatropha curcas] |
| 14 |
Hb_010812_080 |
0.1765817618 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 15 |
Hb_008066_020 |
0.1772418968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000087_110 |
0.1772628321 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005081_050 |
0.1772710389 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor WER-like [Prunus mume] |
| 18 |
Hb_093541_010 |
0.177307605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_073680_010 |
0.1773172016 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 20 |
Hb_157950_010 |
0.1773206197 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Jatropha curcas] |