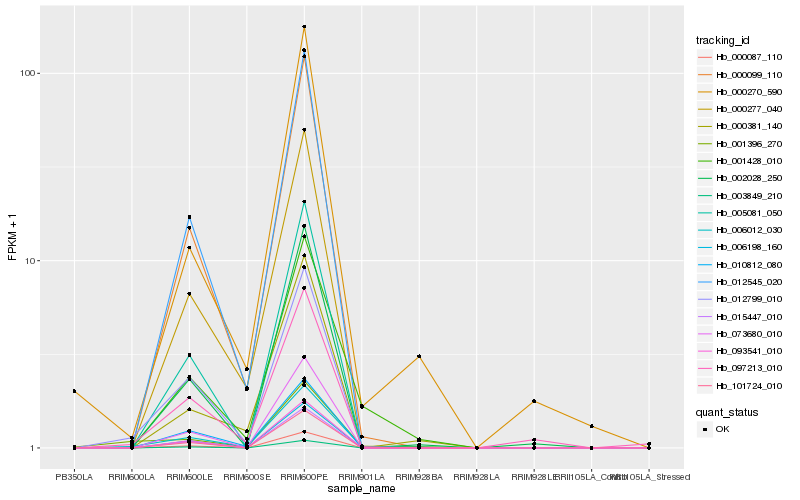
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006012_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s02380g [Populus trichocarpa] |
| 2 |
Hb_012799_010 |
0.1053263692 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000099_110 |
0.1161628666 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 4 |
Hb_012545_020 |
0.1246336005 |
- |
- |
PREDICTED: uncharacterized protein LOC105633903 [Jatropha curcas] |
| 5 |
Hb_001396_270 |
0.1251977084 |
- |
- |
4-coumarate--CoA ligase family protein [Populus trichocarpa] |
| 6 |
Hb_001428_010 |
0.1275746644 |
- |
- |
PREDICTED: transcription factor PRE1-like [Jatropha curcas] |
| 7 |
Hb_000277_040 |
0.1296170721 |
- |
- |
- |
| 8 |
Hb_000270_590 |
0.1318138182 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 9 |
Hb_097213_010 |
0.1348256575 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Populus euphratica] |
| 10 |
Hb_010812_080 |
0.1356924739 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 11 |
Hb_000381_140 |
0.1364336172 |
- |
- |
PREDICTED: S-type anion channel SLAH4-like [Jatropha curcas] |
| 12 |
Hb_002028_250 |
0.1401696589 |
- |
- |
Alpha-aminoadipic semialdehyde synthase, mitochondrial [Crassostrea gigas] |
| 13 |
Hb_101724_010 |
0.1413998046 |
- |
- |
glutamate receptor family protein [Populus trichocarpa] |
| 14 |
Hb_006198_160 |
0.1414184318 |
- |
- |
hypothetical protein POPTR_0002s24290g [Populus trichocarpa] |
| 15 |
Hb_093541_010 |
0.1414189626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000087_110 |
0.1414754384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_015447_010 |
0.1414834837 |
- |
- |
PREDICTED: uncharacterized protein LOC105111046 [Populus euphratica] |
| 18 |
Hb_003849_210 |
0.1418516706 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0793640 [Ricinus communis] |
| 19 |
Hb_005081_050 |
0.1418787768 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor WER-like [Prunus mume] |
| 20 |
Hb_073680_010 |
0.142043625 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |