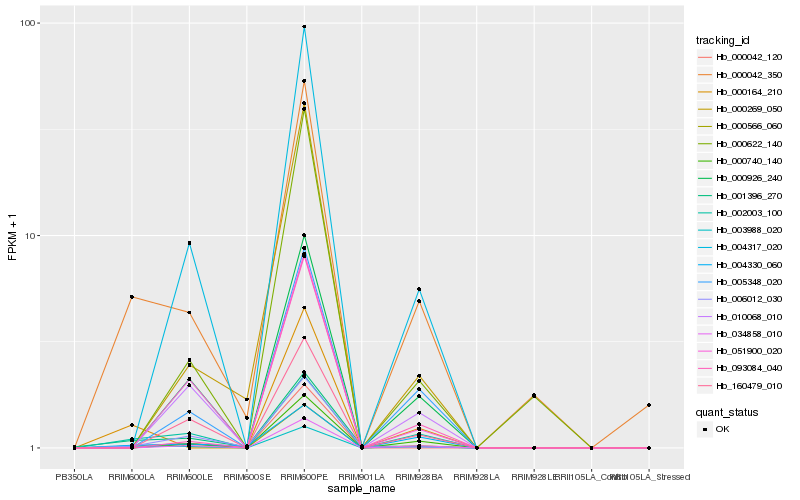
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003988_020 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH85-like [Populus euphratica] |
| 2 |
Hb_000042_350 |
0.1186391694 |
- |
- |
hypothetical protein JCGZ_00710 [Jatropha curcas] |
| 3 |
Hb_002003_100 |
0.1279750717 |
- |
- |
- |
| 4 |
Hb_001396_270 |
0.1597239456 |
- |
- |
4-coumarate--CoA ligase family protein [Populus trichocarpa] |
| 5 |
Hb_000164_210 |
0.160585412 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 6 |
Hb_034858_010 |
0.1861861873 |
- |
- |
- |
| 7 |
Hb_000042_120 |
0.1896953858 |
- |
- |
PREDICTED: uncharacterized protein LOC105632800 [Jatropha curcas] |
| 8 |
Hb_004317_020 |
0.1901798632 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7-like [Malus domestica] |
| 9 |
Hb_000566_060 |
0.1923651965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000740_140 |
0.1939825144 |
transcription factor |
TF Family: OFP |
PREDICTED: probable transcription repressor OFP9 [Jatropha curcas] |
| 11 |
Hb_004330_060 |
0.1969321324 |
- |
- |
hypothetical protein POPTR_0011s01530g [Populus trichocarpa] |
| 12 |
Hb_005348_020 |
0.1993974465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000926_240 |
0.2033785897 |
- |
- |
hypothetical protein PRUPE_ppb018041mg [Prunus persica] |
| 14 |
Hb_006012_030 |
0.2048019324 |
- |
- |
hypothetical protein POPTR_0008s02380g [Populus trichocarpa] |
| 15 |
Hb_010068_010 |
0.2065819226 |
- |
- |
PREDICTED: uncharacterized protein LOC105633273 [Jatropha curcas] |
| 16 |
Hb_000269_050 |
0.2162803128 |
- |
- |
PREDICTED: beta-glucosidase 12-like [Gossypium raimondii] |
| 17 |
Hb_160479_010 |
0.216543943 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 18 |
Hb_000622_140 |
0.2175517891 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_093084_040 |
0.2175759281 |
transcription factor |
TF Family: LOB |
hypothetical protein POPTR_0015s09370g [Populus trichocarpa] |
| 20 |
Hb_051900_020 |
0.2212472106 |
- |
- |
JHL22C18.7 [Jatropha curcas] |