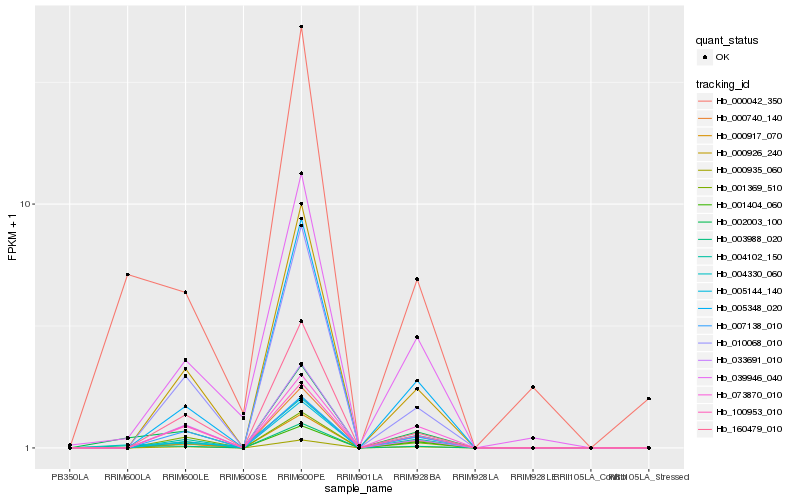
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002003_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_003988_020 |
0.1279750717 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH85-like [Populus euphratica] |
| 3 |
Hb_004330_060 |
0.1575388042 |
- |
- |
hypothetical protein POPTR_0011s01530g [Populus trichocarpa] |
| 4 |
Hb_000042_350 |
0.1591399666 |
- |
- |
hypothetical protein JCGZ_00710 [Jatropha curcas] |
| 5 |
Hb_000935_060 |
0.1773077246 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 6 |
Hb_004102_150 |
0.1774830333 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 7 |
Hb_000926_240 |
0.1872364062 |
- |
- |
hypothetical protein PRUPE_ppb018041mg [Prunus persica] |
| 8 |
Hb_000917_070 |
0.1887459085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005144_140 |
0.1899800085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_039946_040 |
0.1901333537 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 11 |
Hb_010068_010 |
0.1955421807 |
- |
- |
PREDICTED: uncharacterized protein LOC105633273 [Jatropha curcas] |
| 12 |
Hb_160479_010 |
0.1962882708 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 13 |
Hb_073870_010 |
0.1982411112 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 14 |
Hb_001369_510 |
0.199958171 |
- |
- |
PREDICTED: uncharacterized protein LOC105766768 [Gossypium raimondii] |
| 15 |
Hb_007138_010 |
0.2006194596 |
- |
- |
PREDICTED: putative laccase-9 [Jatropha curcas] |
| 16 |
Hb_005348_020 |
0.2007707149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_100953_010 |
0.2012495274 |
- |
- |
hypothetical protein JCGZ_10310 [Jatropha curcas] |
| 18 |
Hb_033691_010 |
0.2043189033 |
- |
- |
Lignin-forming anionic peroxidase precursor, putative [Ricinus communis] |
| 19 |
Hb_001404_060 |
0.2085210774 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000740_140 |
0.2100015162 |
transcription factor |
TF Family: OFP |
PREDICTED: probable transcription repressor OFP9 [Jatropha curcas] |