| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
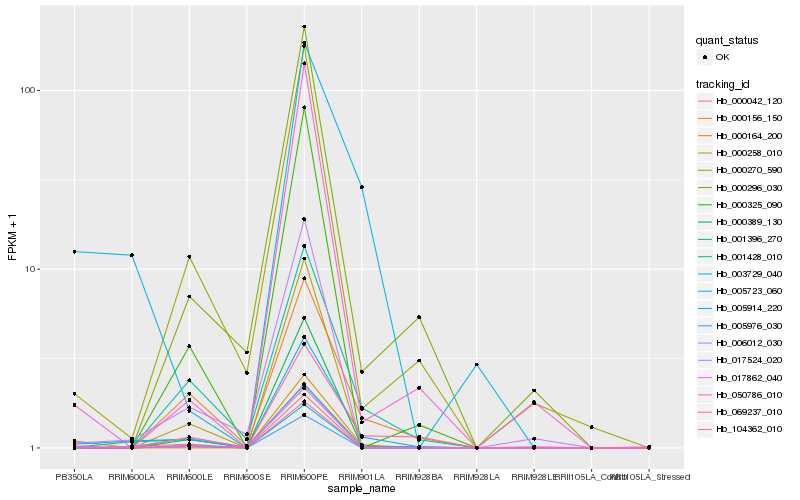
Hb_005914_220 |
0.0 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001396_270 |
0.145017699 |
- |
- |
4-coumarate--CoA ligase family protein [Populus trichocarpa] |
| 3 |
Hb_000042_120 |
0.1770275132 |
- |
- |
PREDICTED: uncharacterized protein LOC105632800 [Jatropha curcas] |
| 4 |
Hb_000156_150 |
0.1808883945 |
- |
- |
hypothetical protein JCGZ_04603 [Jatropha curcas] |
| 5 |
Hb_006012_030 |
0.1868650705 |
- |
- |
hypothetical protein POPTR_0008s02380g [Populus trichocarpa] |
| 6 |
Hb_000389_130 |
0.1891345439 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 7 |
Hb_001428_010 |
0.1895486778 |
- |
- |
PREDICTED: transcription factor PRE1-like [Jatropha curcas] |
| 8 |
Hb_104362_010 |
0.1924962887 |
- |
- |
hypothetical protein POPTR_0247s002302g, partial [Populus trichocarpa] |
| 9 |
Hb_000270_590 |
0.1934588017 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 10 |
Hb_017524_020 |
0.198798239 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 11 |
Hb_017862_040 |
0.2027364972 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 3-like [Prunus mume] |
| 12 |
Hb_000164_200 |
0.2042973792 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 13 |
Hb_005723_060 |
0.2050340975 |
- |
- |
- |
| 14 |
Hb_050786_010 |
0.206076182 |
- |
- |
hypothetical protein MIMGU_mgv1a014824mg [Erythranthe guttata] |
| 15 |
Hb_000296_030 |
0.2061096533 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 16 |
Hb_000325_090 |
0.2061786511 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PRE5-like [Gossypium raimondii] |
| 17 |
Hb_069237_010 |
0.2105142873 |
- |
- |
- |
| 18 |
Hb_000258_010 |
0.2105294042 |
- |
- |
lipid transfer precursor protein [Hevea brasiliensis] |
| 19 |
Hb_005976_030 |
0.2107350192 |
- |
- |
PREDICTED: uncharacterized protein LOC105639715 [Jatropha curcas] |
| 20 |
Hb_003729_040 |
0.2107488163 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |