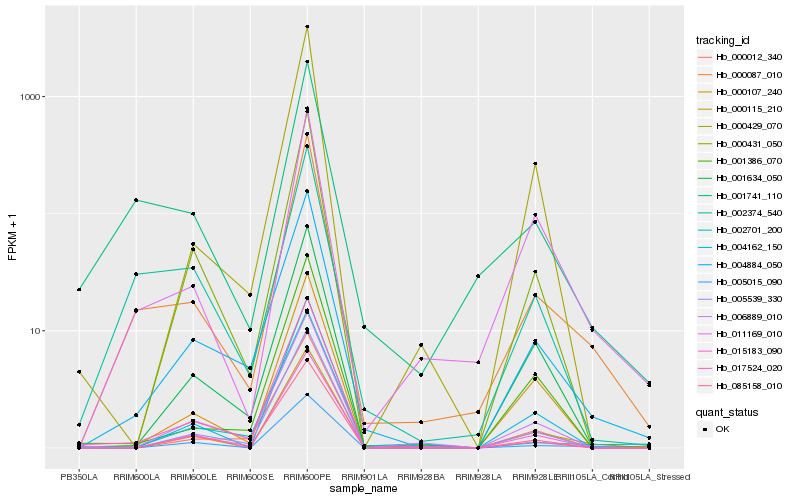
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000087_010 |
0.0 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 2 |
Hb_004884_050 |
0.1184801946 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 3 |
Hb_001741_110 |
0.1193581056 |
rubber biosynthesis |
Gene Name: REF7 |
REF/SRPP-like protein [Glycine soja] |
| 4 |
Hb_011169_010 |
0.129663196 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 5 |
Hb_000115_210 |
0.1379859226 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 3-like [Jatropha curcas] |
| 6 |
Hb_002374_540 |
0.1417764372 |
- |
- |
RecName: Full=Esterase; AltName: Full=Early nodule-specific protein homolog; AltName: Full=Latex allergen Hev b 13; AltName: Allergen=Hev b 13; Flags: Precursor [Hevea brasiliensis] |
| 7 |
Hb_006889_010 |
0.1515461888 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 8 |
Hb_005539_330 |
0.1540240051 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 9 |
Hb_085158_010 |
0.1549352458 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 10 |
Hb_005015_090 |
0.156367432 |
- |
- |
PREDICTED: cytochrome P450 78A7 [Jatropha curcas] |
| 11 |
Hb_000107_240 |
0.15670168 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 12 |
Hb_004162_150 |
0.1567632441 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 13 |
Hb_015183_090 |
0.1570450338 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 14 |
Hb_001634_050 |
0.1574041196 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 15 |
Hb_000431_050 |
0.1574086959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_017524_020 |
0.1582245929 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 17 |
Hb_002701_200 |
0.1613611101 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 18 |
Hb_001386_070 |
0.1614391514 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 19 |
Hb_000429_070 |
0.1641371699 |
- |
- |
hypothetical protein POPTR_0006s05480g [Populus trichocarpa] |
| 20 |
Hb_000012_340 |
0.164489407 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |