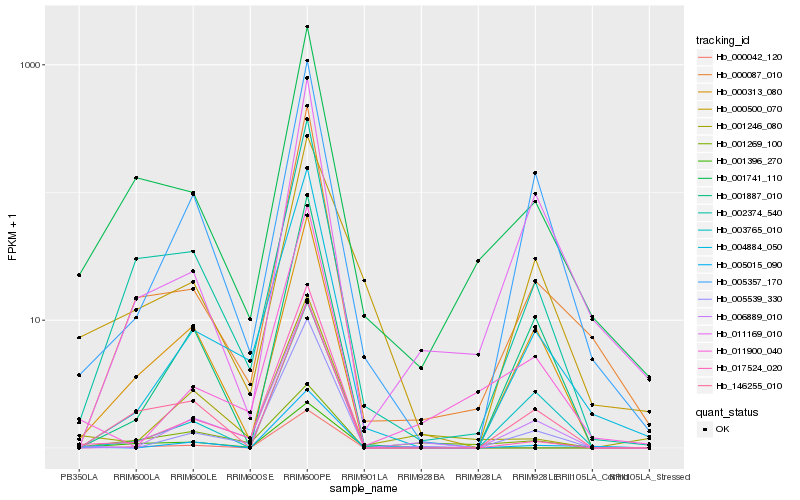
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001741_110 |
0.0 |
rubber biosynthesis |
Gene Name: REF7 |
REF/SRPP-like protein [Glycine soja] |
| 2 |
Hb_002374_540 |
0.1128567039 |
- |
- |
RecName: Full=Esterase; AltName: Full=Early nodule-specific protein homolog; AltName: Full=Latex allergen Hev b 13; AltName: Allergen=Hev b 13; Flags: Precursor [Hevea brasiliensis] |
| 3 |
Hb_000087_010 |
0.1193581056 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 4 |
Hb_146255_010 |
0.159923194 |
- |
- |
PREDICTED: uncharacterized protein LOC105643649 [Jatropha curcas] |
| 5 |
Hb_011169_010 |
0.1804542427 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 6 |
Hb_004884_050 |
0.1842998694 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 7 |
Hb_000313_080 |
0.1849511578 |
rubber biosynthesis |
Gene Name: CPT3 |
cis-prenyltransferase 3 [Hevea brasiliensis] |
| 8 |
Hb_011900_040 |
0.1852331498 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 9 |
Hb_001396_270 |
0.1860192259 |
- |
- |
4-coumarate--CoA ligase family protein [Populus trichocarpa] |
| 10 |
Hb_000500_070 |
0.1895563978 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 11 |
Hb_005357_170 |
0.1990649809 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 12 |
Hb_017524_020 |
0.1995977051 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 13 |
Hb_005539_330 |
0.2030560898 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 14 |
Hb_003765_010 |
0.2050844876 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 15 |
Hb_001887_010 |
0.2075893968 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 16 |
Hb_000042_120 |
0.2076781401 |
- |
- |
PREDICTED: uncharacterized protein LOC105632800 [Jatropha curcas] |
| 17 |
Hb_006889_010 |
0.2077431478 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 18 |
Hb_005015_090 |
0.2077641443 |
- |
- |
PREDICTED: cytochrome P450 78A7 [Jatropha curcas] |
| 19 |
Hb_001246_080 |
0.2080426596 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 20 |
Hb_001269_100 |
0.2088292459 |
- |
- |
plasma membrane ATPase 1, partial [Triticum aestivum] |