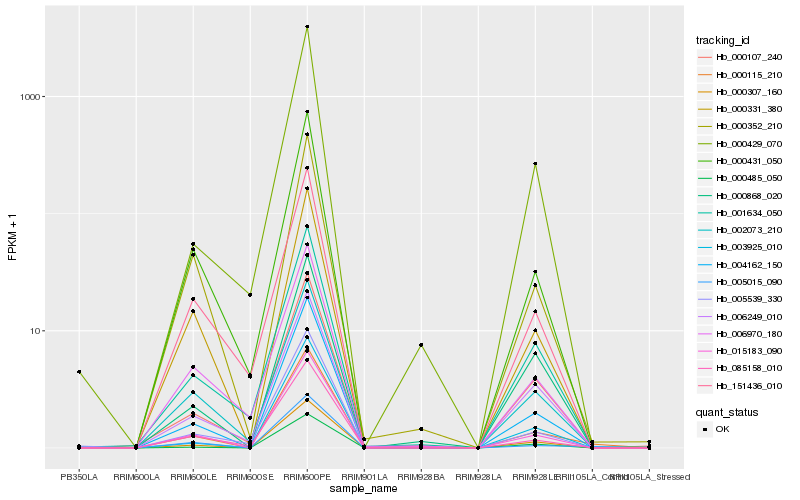
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000115_210 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 3-like [Jatropha curcas] |
| 2 |
Hb_004162_150 |
0.0771063106 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 3 |
Hb_000307_160 |
0.0827402074 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 4 |
Hb_005015_090 |
0.0933619186 |
- |
- |
PREDICTED: cytochrome P450 78A7 [Jatropha curcas] |
| 5 |
Hb_015183_090 |
0.0962835762 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 6 |
Hb_000331_380 |
0.1001531977 |
- |
- |
hypothetical protein POPTR_0012s09560g [Populus trichocarpa] |
| 7 |
Hb_000431_050 |
0.1005885745 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000485_050 |
0.1037627779 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 9 |
Hb_002073_210 |
0.106696753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003925_010 |
0.1084404505 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 11 |
Hb_085158_010 |
0.1096100377 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 12 |
Hb_000352_210 |
0.1097637918 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 13 |
Hb_000107_240 |
0.1101895583 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 14 |
Hb_001634_050 |
0.1113920631 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 15 |
Hb_151436_010 |
0.1164834567 |
- |
- |
PREDICTED: uncharacterized protein LOC105137019 [Populus euphratica] |
| 16 |
Hb_000429_070 |
0.1186631468 |
- |
- |
hypothetical protein POPTR_0006s05480g [Populus trichocarpa] |
| 17 |
Hb_005539_330 |
0.1194922297 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 18 |
Hb_006249_010 |
0.1211234245 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 19 |
Hb_000868_020 |
0.1231208858 |
- |
- |
- |
| 20 |
Hb_006970_180 |
0.1240965176 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |