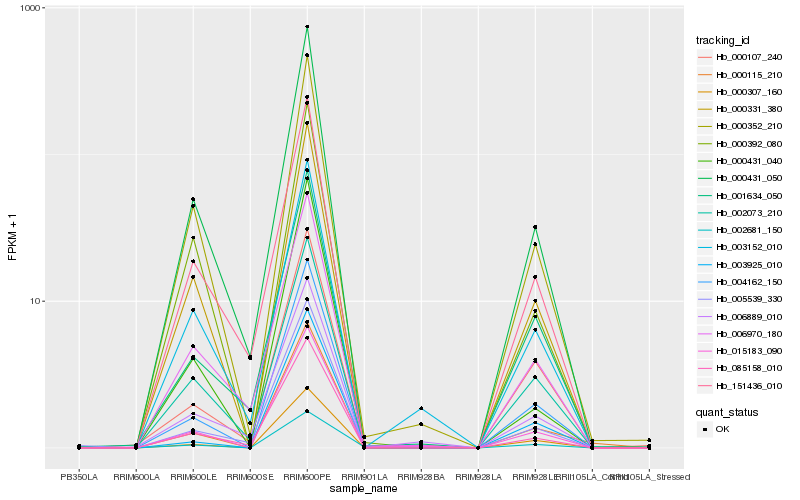
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015183_090 |
0.0 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 2 |
Hb_004162_150 |
0.0666591774 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 3 |
Hb_000352_210 |
0.0735544881 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 4 |
Hb_000331_380 |
0.0748418408 |
- |
- |
hypothetical protein POPTR_0012s09560g [Populus trichocarpa] |
| 5 |
Hb_000431_050 |
0.0768687401 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000307_160 |
0.0790033355 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 7 |
Hb_003152_010 |
0.0793428898 |
- |
- |
- |
| 8 |
Hb_006889_010 |
0.0893674035 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 9 |
Hb_085158_010 |
0.0894125446 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 10 |
Hb_002073_210 |
0.0909435897 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005539_330 |
0.0922659069 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 12 |
Hb_002681_150 |
0.0924977369 |
- |
- |
uridine cytidine kinase I, putative [Ricinus communis] |
| 13 |
Hb_006970_180 |
0.0962025316 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 14 |
Hb_000115_210 |
0.0962835762 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 3-like [Jatropha curcas] |
| 15 |
Hb_151436_010 |
0.0986253835 |
- |
- |
PREDICTED: uncharacterized protein LOC105137019 [Populus euphratica] |
| 16 |
Hb_000107_240 |
0.1006253054 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 17 |
Hb_001634_050 |
0.1016290314 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 18 |
Hb_003925_010 |
0.1031612686 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 19 |
Hb_000431_040 |
0.1052310019 |
- |
- |
PREDICTED: uncharacterized protein LOC105635551 [Jatropha curcas] |
| 20 |
Hb_000392_080 |
0.1082025636 |
- |
- |
hypothetical protein glysoja_034865 [Glycine soja] |