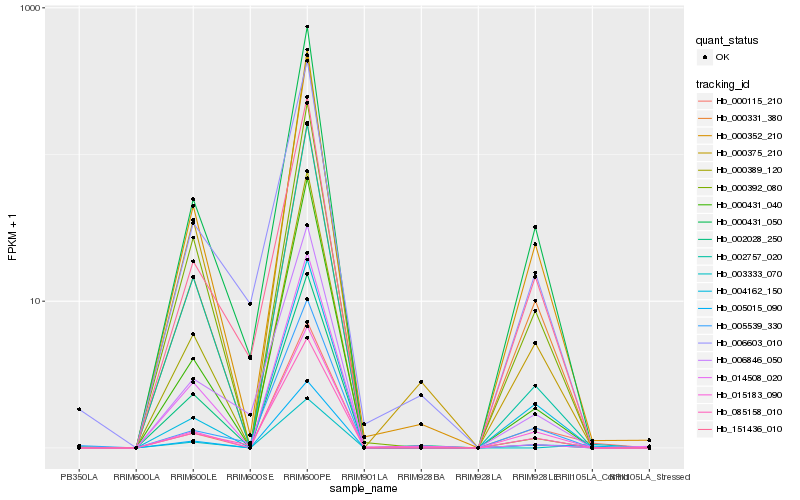
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005015_090 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 78A7 [Jatropha curcas] |
| 2 |
Hb_000115_210 |
0.0933619186 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 3-like [Jatropha curcas] |
| 3 |
Hb_000431_040 |
0.1106576118 |
- |
- |
PREDICTED: uncharacterized protein LOC105635551 [Jatropha curcas] |
| 4 |
Hb_000431_050 |
0.1151052455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002757_020 |
0.1157746013 |
- |
- |
- |
| 6 |
Hb_000352_210 |
0.1189513711 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 7 |
Hb_000331_380 |
0.1203549465 |
- |
- |
hypothetical protein POPTR_0012s09560g [Populus trichocarpa] |
| 8 |
Hb_085158_010 |
0.1210835052 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 9 |
Hb_000375_210 |
0.1218852982 |
- |
- |
- |
| 10 |
Hb_000392_080 |
0.1247509414 |
- |
- |
hypothetical protein glysoja_034865 [Glycine soja] |
| 11 |
Hb_015183_090 |
0.1273519074 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 12 |
Hb_005539_330 |
0.1278375123 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 13 |
Hb_004162_150 |
0.1282507501 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 14 |
Hb_000389_120 |
0.1333955454 |
- |
- |
hypothetical protein JCGZ_20066 [Jatropha curcas] |
| 15 |
Hb_003333_070 |
0.1354287165 |
- |
- |
Ent-kaurenoic acid oxidase, putative [Ricinus communis] |
| 16 |
Hb_006603_010 |
0.1383636744 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 17 |
Hb_151436_010 |
0.1384256162 |
- |
- |
PREDICTED: uncharacterized protein LOC105137019 [Populus euphratica] |
| 18 |
Hb_002028_250 |
0.1391365124 |
- |
- |
Alpha-aminoadipic semialdehyde synthase, mitochondrial [Crassostrea gigas] |
| 19 |
Hb_014508_020 |
0.1404173977 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006846_050 |
0.1421182597 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |