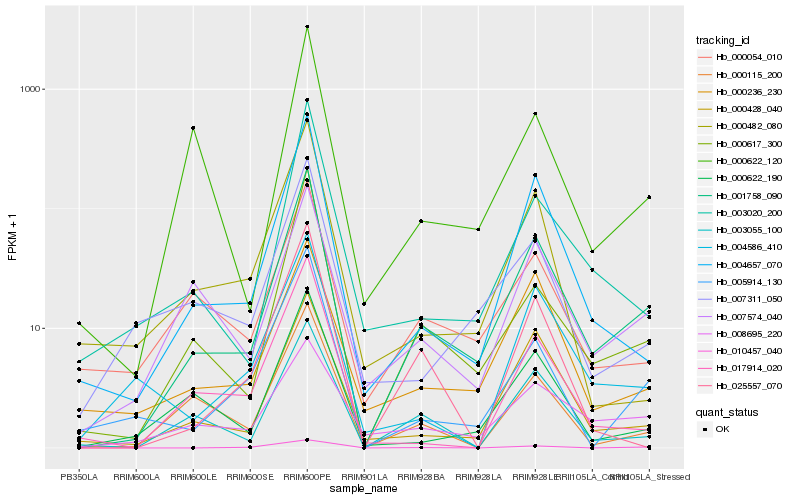
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001758_090 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 2 |
Hb_004657_070 |
0.1491647855 |
- |
- |
PREDICTED: protein IRX15-LIKE [Jatropha curcas] |
| 3 |
Hb_000617_300 |
0.1527298834 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 4 |
Hb_000428_040 |
0.1570289124 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Populus euphratica] |
| 5 |
Hb_003055_100 |
0.1654477718 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 6 |
Hb_007574_040 |
0.1715737745 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 7 |
Hb_000622_190 |
0.1763443426 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |
| 8 |
Hb_003020_200 |
0.1780733633 |
- |
- |
PREDICTED: uncharacterized protein LOC105131117 [Populus euphratica] |
| 9 |
Hb_000236_230 |
0.1815082118 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 10 |
Hb_010457_040 |
0.1825094579 |
- |
- |
PREDICTED: uncharacterized protein LOC101491058 [Cicer arietinum] |
| 11 |
Hb_000115_200 |
0.1835534805 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 12 |
Hb_005914_130 |
0.1838383114 |
- |
- |
- |
| 13 |
Hb_008695_220 |
0.1852231798 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 14 |
Hb_000054_010 |
0.1887554149 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007311_050 |
0.190918759 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004586_410 |
0.1913686435 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 17 |
Hb_000622_120 |
0.1939424277 |
- |
- |
hypothetical protein [Hevea brasiliensis] |
| 18 |
Hb_000482_080 |
0.1952711293 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 19 |
Hb_017914_020 |
0.1969935304 |
- |
- |
- |
| 20 |
Hb_025557_070 |
0.197756416 |
transcription factor |
TF Family: zf-HD |
hypothetical protein JCGZ_11626 [Jatropha curcas] |