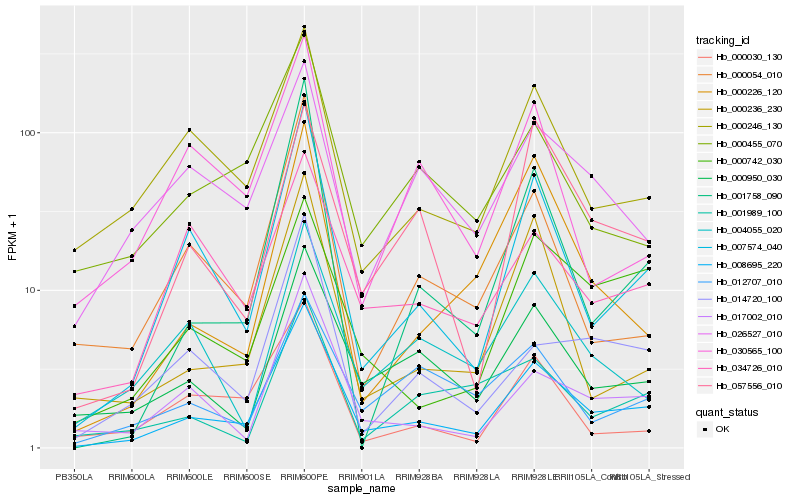
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008695_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 2 |
Hb_007574_040 |
0.1363339255 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 3 |
Hb_000950_030 |
0.1583575897 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 4 |
Hb_017002_010 |
0.1618308568 |
- |
- |
PREDICTED: expansin-A15-like [Jatropha curcas] |
| 5 |
Hb_000246_130 |
0.1656838301 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 6 |
Hb_000455_070 |
0.1773799824 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 7 |
Hb_014720_100 |
0.1796857317 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 8 |
Hb_000236_230 |
0.1821615842 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000742_030 |
0.1822602517 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 10 |
Hb_000054_010 |
0.1831754893 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001758_090 |
0.1852231798 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 12 |
Hb_034726_010 |
0.1889660618 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X2 [Jatropha curcas] |
| 13 |
Hb_000030_130 |
0.1898790248 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_057556_010 |
0.1929262913 |
- |
- |
Light-regulated protein precursor, putative [Ricinus communis] |
| 15 |
Hb_004055_020 |
0.1953357542 |
- |
- |
PREDICTED: uncharacterized protein LOC105631433 [Jatropha curcas] |
| 16 |
Hb_026527_010 |
0.1961316077 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 17 |
Hb_000226_120 |
0.1961733837 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_012707_010 |
0.1967588878 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 19 |
Hb_030565_100 |
0.1995573227 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 20 |
Hb_001989_100 |
0.2022010375 |
- |
- |
PREDICTED: uncharacterized protein LOC105636214 [Jatropha curcas] |