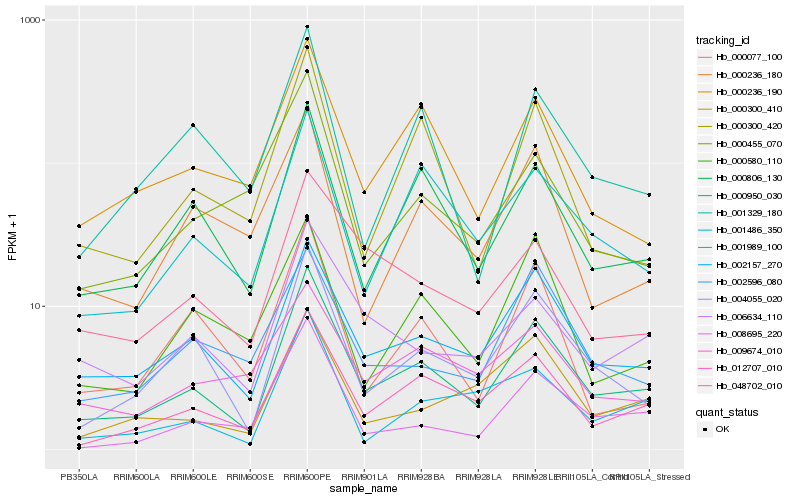
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000950_030 |
0.0 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 2 |
Hb_002157_270 |
0.1143793085 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 3 |
Hb_012707_010 |
0.116475251 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 4 |
Hb_004055_020 |
0.1275555277 |
- |
- |
PREDICTED: uncharacterized protein LOC105631433 [Jatropha curcas] |
| 5 |
Hb_000806_130 |
0.1378379033 |
- |
- |
Adenosine kinase 2 [Theobroma cacao] |
| 6 |
Hb_001486_350 |
0.156509883 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_006634_110 |
0.1579592977 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 8 |
Hb_008695_220 |
0.1583575897 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 9 |
Hb_000236_190 |
0.159863938 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 10 |
Hb_000300_420 |
0.165676738 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_048702_010 |
0.1660895901 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_001989_100 |
0.1683161068 |
- |
- |
PREDICTED: uncharacterized protein LOC105636214 [Jatropha curcas] |
| 13 |
Hb_001329_180 |
0.1691619879 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 14 |
Hb_000300_410 |
0.1702336576 |
- |
- |
hypothetical protein POPTR_0013s05860g [Populus trichocarpa] |
| 15 |
Hb_000077_100 |
0.174408281 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000580_110 |
0.1798671291 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 17 |
Hb_000236_180 |
0.180016913 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000455_070 |
0.1804923565 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 19 |
Hb_009674_010 |
0.1811689722 |
- |
- |
AAEL007687-PA [Aedes aegypti] |
| 20 |
Hb_002596_080 |
0.181407062 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |