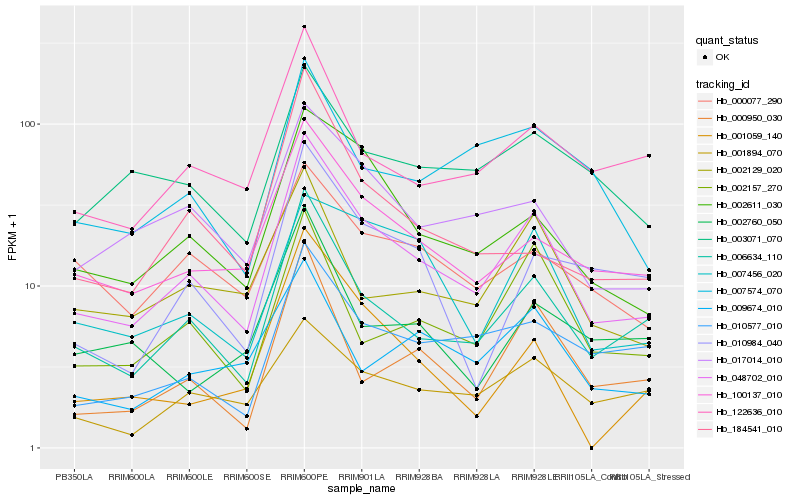
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048702_010 |
0.0 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_006634_110 |
0.1022030405 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 3 |
Hb_100137_010 |
0.1275119634 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 4 |
Hb_002611_030 |
0.1347267388 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 5 |
Hb_017014_010 |
0.140106602 |
- |
- |
PREDICTED: syntaxin-132-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_122636_010 |
0.1531455299 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 7 |
Hb_010577_010 |
0.160260349 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase-like [Populus euphratica] |
| 8 |
Hb_002129_020 |
0.1605157846 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 9 |
Hb_002157_270 |
0.1629852057 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 10 |
Hb_000950_030 |
0.1660895901 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 11 |
Hb_001894_070 |
0.1693841766 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003071_070 |
0.1704150679 |
- |
- |
sucrose synthase 3 [Hevea brasiliensis] |
| 13 |
Hb_000077_290 |
0.1705585316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007574_070 |
0.1710808149 |
- |
- |
fatty acid desaturase, putative [Ricinus communis] |
| 15 |
Hb_009674_010 |
0.1722416167 |
- |
- |
AAEL007687-PA [Aedes aegypti] |
| 16 |
Hb_010984_040 |
0.1735545724 |
- |
- |
hypothetical protein M569_17286, partial [Genlisea aurea] |
| 17 |
Hb_184541_010 |
0.1749580398 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 18 |
Hb_007456_020 |
0.1779175833 |
- |
- |
unknown [Lotus japonicus] |
| 19 |
Hb_001059_140 |
0.1837252501 |
- |
- |
hypothetical protein POPTR_0004s03390g [Populus trichocarpa] |
| 20 |
Hb_002760_050 |
0.1838190302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |