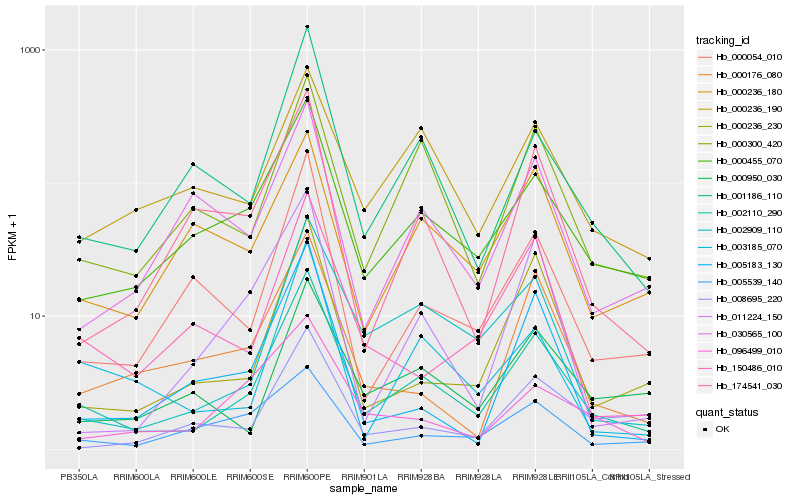
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002110_290 |
0.0 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 2 |
Hb_000455_070 |
0.1453334428 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 3 |
Hb_001186_110 |
0.1670331305 |
- |
- |
hypothetical protein JCGZ_18389 [Jatropha curcas] |
| 4 |
Hb_000300_420 |
0.1725559047 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_002909_110 |
0.1739223754 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 6 |
Hb_003185_070 |
0.174131443 |
- |
- |
Plant natriuretic peptide A [Theobroma cacao] |
| 7 |
Hb_000054_010 |
0.1795539342 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000236_230 |
0.181081768 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 9 |
Hb_174541_030 |
0.1915072322 |
- |
- |
Cytochrome P450, family 98, subfamily A, polypeptide 3 [Theobroma cacao] |
| 10 |
Hb_000950_030 |
0.1923427182 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 11 |
Hb_150486_010 |
0.1953676548 |
- |
- |
hypothetical protein CICLE_v100116733mg, partial [Citrus clementina] |
| 12 |
Hb_000236_190 |
0.1965465323 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 13 |
Hb_000176_080 |
0.1980828223 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g30520 [Jatropha curcas] |
| 14 |
Hb_011224_150 |
0.1995600449 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 15 |
Hb_096499_010 |
0.2020537301 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 16 |
Hb_005183_130 |
0.2032150857 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 17 |
Hb_005539_140 |
0.2076206805 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 18 |
Hb_000236_180 |
0.2080243261 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 19 |
Hb_030565_100 |
0.2093342016 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 20 |
Hb_008695_220 |
0.2112123244 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |