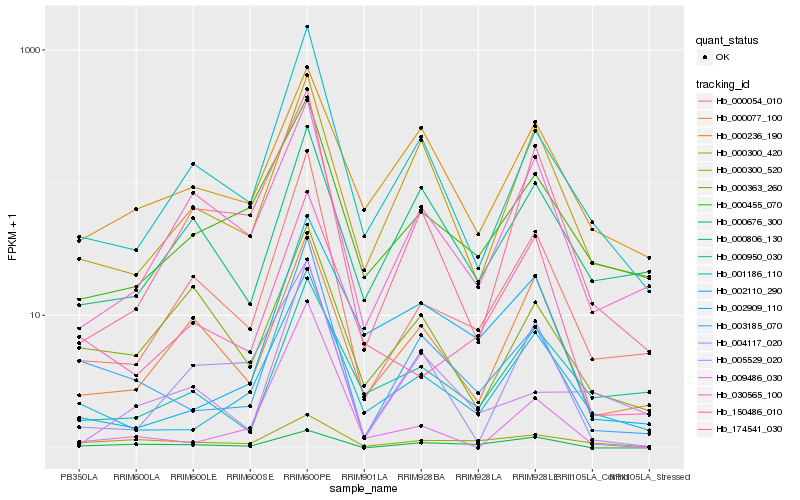
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003185_070 |
0.0 |
- |
- |
Plant natriuretic peptide A [Theobroma cacao] |
| 2 |
Hb_002110_290 |
0.174131443 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 3 |
Hb_001186_110 |
0.1872954341 |
- |
- |
hypothetical protein JCGZ_18389 [Jatropha curcas] |
| 4 |
Hb_000054_010 |
0.2094221039 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000300_420 |
0.2123376421 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000455_070 |
0.2330567896 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 7 |
Hb_000236_190 |
0.2347188444 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 8 |
Hb_000300_520 |
0.2440838368 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 9 |
Hb_000077_100 |
0.2463037994 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_002909_110 |
0.2479527895 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 11 |
Hb_009486_030 |
0.2492188714 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 12 [Jatropha curcas] |
| 12 |
Hb_000676_300 |
0.25202563 |
- |
- |
PREDICTED: probable caffeine synthase 4 [Jatropha curcas] |
| 13 |
Hb_005529_020 |
0.2528399875 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 14 |
Hb_030565_100 |
0.2528462044 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 15 |
Hb_174541_030 |
0.2537622718 |
- |
- |
Cytochrome P450, family 98, subfamily A, polypeptide 3 [Theobroma cacao] |
| 16 |
Hb_004117_020 |
0.2545613099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_150486_010 |
0.2570553652 |
- |
- |
hypothetical protein CICLE_v100116733mg, partial [Citrus clementina] |
| 18 |
Hb_000806_130 |
0.257837291 |
- |
- |
Adenosine kinase 2 [Theobroma cacao] |
| 19 |
Hb_000363_260 |
0.2585096714 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 20 |
Hb_000950_030 |
0.2592466099 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |