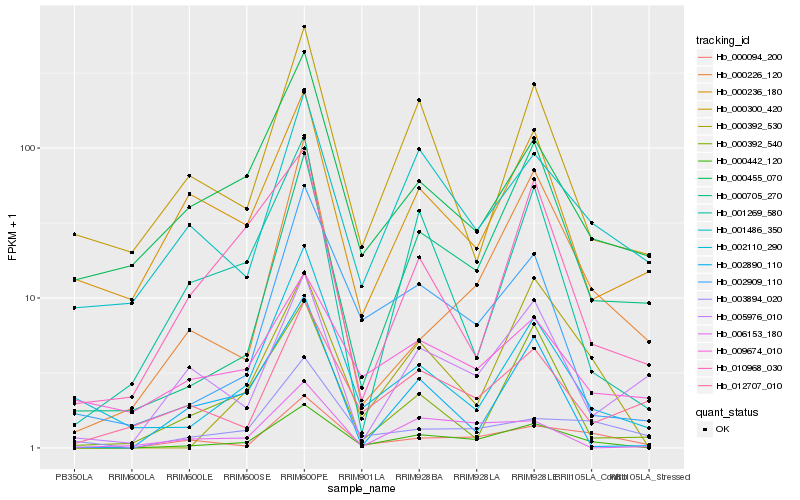
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000442_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000392_530 |
0.1762127647 |
- |
- |
hypothetical protein PHAVU_003G0695001g, partial [Phaseolus vulgaris] |
| 3 |
Hb_002909_110 |
0.1931192995 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 4 |
Hb_000094_200 |
0.1997523896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001486_350 |
0.2216085836 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_001269_580 |
0.2216395044 |
- |
- |
PREDICTED: shikimate O-hydroxycinnamoyltransferase [Jatropha curcas] |
| 7 |
Hb_002110_290 |
0.2219578902 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_002890_110 |
0.2272425129 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 9 |
Hb_000392_540 |
0.2337334603 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Sesamum indicum] |
| 10 |
Hb_000300_420 |
0.2345507161 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000226_120 |
0.2350209777 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000455_070 |
0.2354593487 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 13 |
Hb_006153_180 |
0.2368321879 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_012707_010 |
0.2373260712 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 15 |
Hb_010968_030 |
0.2402850423 |
- |
- |
PREDICTED: uncharacterized protein LOC105643149 [Jatropha curcas] |
| 16 |
Hb_000705_270 |
0.241264226 |
- |
- |
PREDICTED: uncharacterized protein LOC105131117 [Populus euphratica] |
| 17 |
Hb_003894_020 |
0.2457548927 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 18 |
Hb_009674_010 |
0.2472945481 |
- |
- |
AAEL007687-PA [Aedes aegypti] |
| 19 |
Hb_000236_180 |
0.2483407638 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 20 |
Hb_005976_010 |
0.2494258683 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |