| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
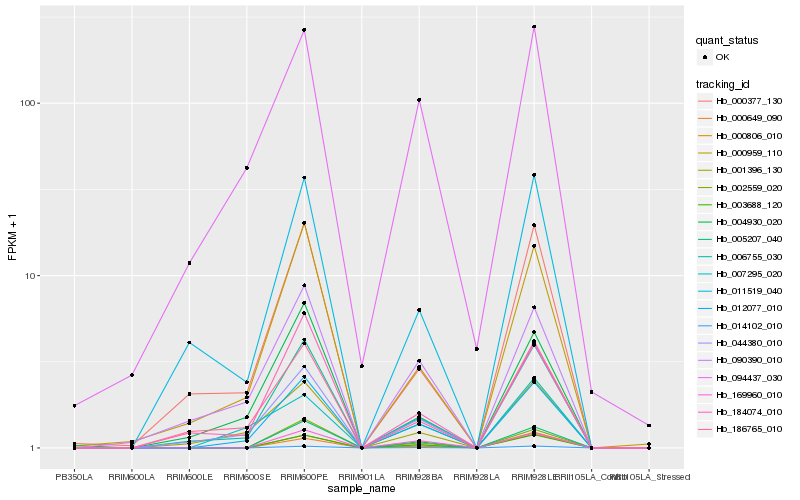
Hb_012077_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 2 |
Hb_006755_030 |
0.1201037416 |
- |
- |
BURP domain-containing protein, putative [Theobroma cacao] |
| 3 |
Hb_003688_120 |
0.1260600915 |
- |
- |
PREDICTED: uncharacterized protein LOC105350793 [Fragaria vesca subsp. vesca] |
| 4 |
Hb_005207_040 |
0.1273690471 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2-like [Cicer arietinum] |
| 5 |
Hb_000959_110 |
0.1290412952 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 6 |
Hb_014102_010 |
0.1329818976 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_001396_130 |
0.1360807391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_169960_010 |
0.1416549088 |
- |
- |
- |
| 9 |
Hb_090390_010 |
0.1443064024 |
- |
- |
hypothetical protein POPTR_0006s28850g [Populus trichocarpa] |
| 10 |
Hb_044380_010 |
0.1446356652 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Malus domestica] |
| 11 |
Hb_002559_020 |
0.1491894076 |
transcription factor |
TF Family: TUB |
- |
| 12 |
Hb_094437_030 |
0.1494019978 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 13 |
Hb_184074_010 |
0.1513079515 |
- |
- |
PREDICTED: uncharacterized protein LOC105639054 [Jatropha curcas] |
| 14 |
Hb_004930_020 |
0.1541787688 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 15 |
Hb_186765_010 |
0.1559043739 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 16 |
Hb_000377_130 |
0.1560876322 |
- |
- |
hypothetical protein POPTR_0016s00790g [Populus trichocarpa] |
| 17 |
Hb_000649_090 |
0.1578434428 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_011519_040 |
0.1585807858 |
- |
- |
hypothetical protein POPTR_0015s04770g [Populus trichocarpa] |
| 19 |
Hb_007295_020 |
0.1629816148 |
- |
- |
Uncharacterized protein TCM_034658 [Theobroma cacao] |
| 20 |
Hb_000806_010 |
0.1668752113 |
- |
- |
PREDICTED: hevamine-A-like [Jatropha curcas] |