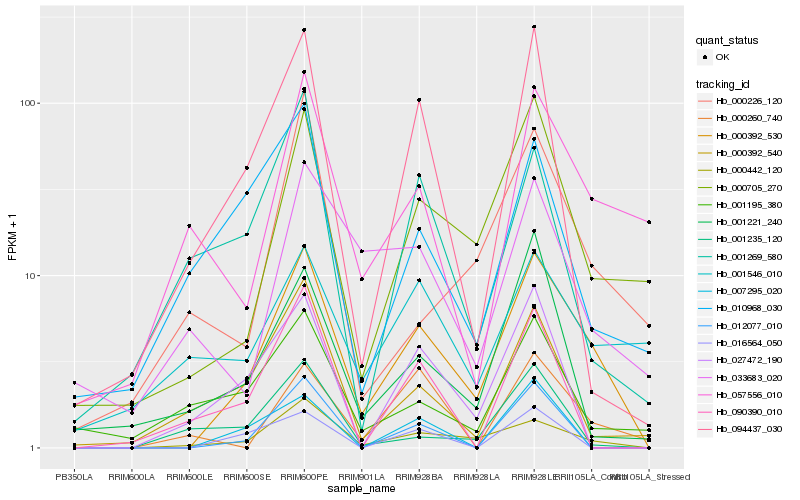
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_530 |
0.0 |
- |
- |
hypothetical protein PHAVU_003G0695001g, partial [Phaseolus vulgaris] |
| 2 |
Hb_000442_120 |
0.1762127647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000705_270 |
0.2075061305 |
- |
- |
PREDICTED: uncharacterized protein LOC105131117 [Populus euphratica] |
| 4 |
Hb_094437_030 |
0.2442856639 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 5 |
Hb_057556_010 |
0.2566761008 |
- |
- |
Light-regulated protein precursor, putative [Ricinus communis] |
| 6 |
Hb_012077_010 |
0.2574969276 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 7 |
Hb_000392_540 |
0.2576296955 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Sesamum indicum] |
| 8 |
Hb_027472_190 |
0.258321205 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 9 |
Hb_001221_240 |
0.2613781382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000226_120 |
0.2684932881 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_010968_030 |
0.2759207696 |
- |
- |
PREDICTED: uncharacterized protein LOC105643149 [Jatropha curcas] |
| 12 |
Hb_033683_020 |
0.2760907291 |
- |
- |
hypothetical protein POPTR_1072s00200g [Populus trichocarpa] |
| 13 |
Hb_001195_380 |
0.2779038026 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 14 |
Hb_001235_120 |
0.2779440879 |
- |
- |
hypothetical protein JCGZ_03395 [Jatropha curcas] |
| 15 |
Hb_007295_020 |
0.2819241661 |
- |
- |
Uncharacterized protein TCM_034658 [Theobroma cacao] |
| 16 |
Hb_000260_740 |
0.2820947547 |
- |
- |
- |
| 17 |
Hb_016564_050 |
0.282754999 |
- |
- |
- |
| 18 |
Hb_001269_580 |
0.2832706397 |
- |
- |
PREDICTED: shikimate O-hydroxycinnamoyltransferase [Jatropha curcas] |
| 19 |
Hb_090390_010 |
0.2838685146 |
- |
- |
hypothetical protein POPTR_0006s28850g [Populus trichocarpa] |
| 20 |
Hb_001546_010 |
0.2841015833 |
- |
- |
conserved hypothetical protein [Ricinus communis] |