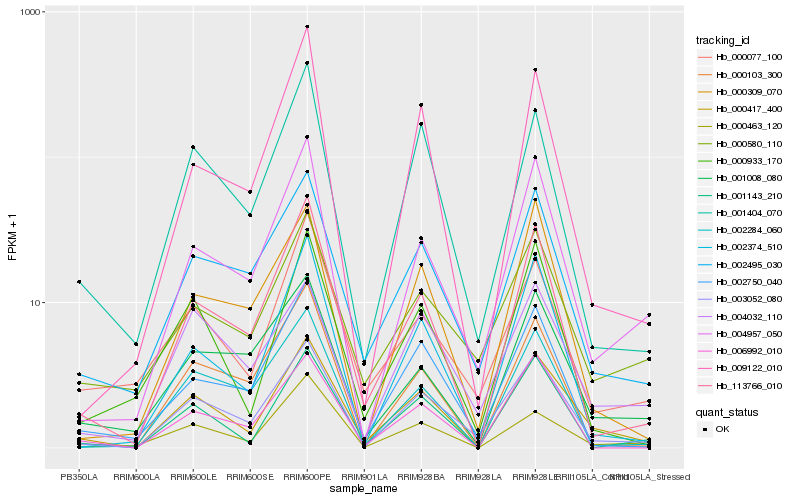
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_400 |
0.0 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_003052_080 |
0.132222935 |
desease resistance |
Gene Name: PRK |
PREDICTED: uridine-cytidine kinase C [Jatropha curcas] |
| 3 |
Hb_113766_010 |
0.1417706525 |
- |
- |
plasma membrane H+ ATPase, partial [Nicotiana plumbaginifolia] |
| 4 |
Hb_001143_210 |
0.1461002892 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 5 |
Hb_002495_030 |
0.1531046814 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001404_070 |
0.1544152862 |
- |
- |
sucrose synthase 4 [Hevea brasiliensis] |
| 7 |
Hb_000933_170 |
0.1572265469 |
- |
- |
PREDICTED: uncharacterized protein At5g01610 [Jatropha curcas] |
| 8 |
Hb_000463_120 |
0.1626954068 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 9 |
Hb_004957_050 |
0.1637263784 |
- |
- |
hypothetical protein POPTR_0010s12360g [Populus trichocarpa] |
| 10 |
Hb_002374_510 |
0.1658829604 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 11 |
Hb_006992_010 |
0.1678031854 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 12 |
Hb_001008_080 |
0.1720294095 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 13 |
Hb_009122_010 |
0.1723502873 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_002750_040 |
0.1729459908 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 15 |
Hb_000077_100 |
0.1781200608 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_004032_110 |
0.1783013138 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000309_070 |
0.1789515413 |
- |
- |
PREDICTED: 2-methylene-furan-3-one reductase [Cucumis melo] |
| 18 |
Hb_000103_300 |
0.179496236 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 19 |
Hb_002284_060 |
0.1804672919 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 20 |
Hb_000580_110 |
0.1808229942 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |