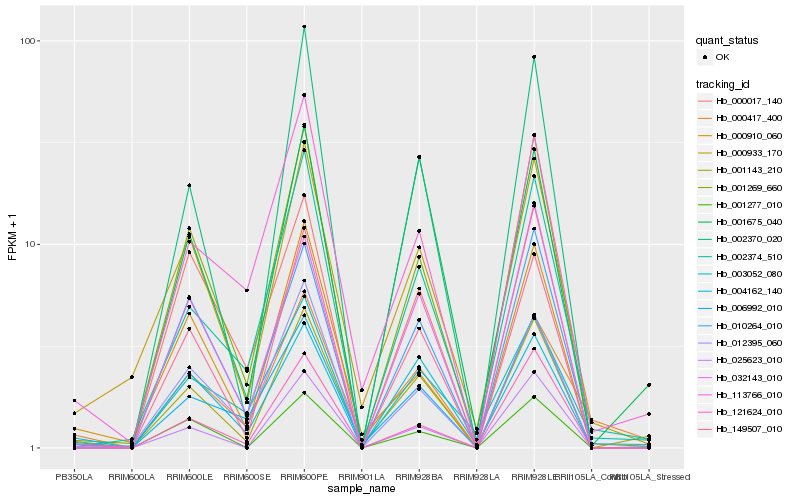
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001143_210 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 2 |
Hb_149507_010 |
0.122106837 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Malus domestica] |
| 3 |
Hb_001269_660 |
0.1260679725 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 4 |
Hb_010264_010 |
0.1372150335 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR-LIKE 1-like [Jatropha curcas] |
| 5 |
Hb_006992_010 |
0.138815048 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 6 |
Hb_000933_170 |
0.1395204862 |
- |
- |
PREDICTED: uncharacterized protein At5g01610 [Jatropha curcas] |
| 7 |
Hb_002370_020 |
0.1397642238 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 8 |
Hb_002374_510 |
0.141918836 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 9 |
Hb_113766_010 |
0.1425193335 |
- |
- |
plasma membrane H+ ATPase, partial [Nicotiana plumbaginifolia] |
| 10 |
Hb_000417_400 |
0.1461002892 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_121624_010 |
0.1506301142 |
- |
- |
PREDICTED: uncharacterized protein LOC105634410 [Jatropha curcas] |
| 12 |
Hb_012395_060 |
0.1527663812 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 13 |
Hb_004162_140 |
0.1531972167 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 14 |
Hb_001675_040 |
0.1538273294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_025623_010 |
0.1560548688 |
- |
- |
- |
| 16 |
Hb_000910_060 |
0.1574915956 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 17 |
Hb_003052_080 |
0.1576358698 |
desease resistance |
Gene Name: PRK |
PREDICTED: uridine-cytidine kinase C [Jatropha curcas] |
| 18 |
Hb_032143_010 |
0.1581617824 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 19 |
Hb_001277_010 |
0.1607010633 |
- |
- |
unknown [Populus trichocarpa] |
| 20 |
Hb_000017_140 |
0.1651812603 |
- |
- |
polygalacturonase, putative [Ricinus communis] |