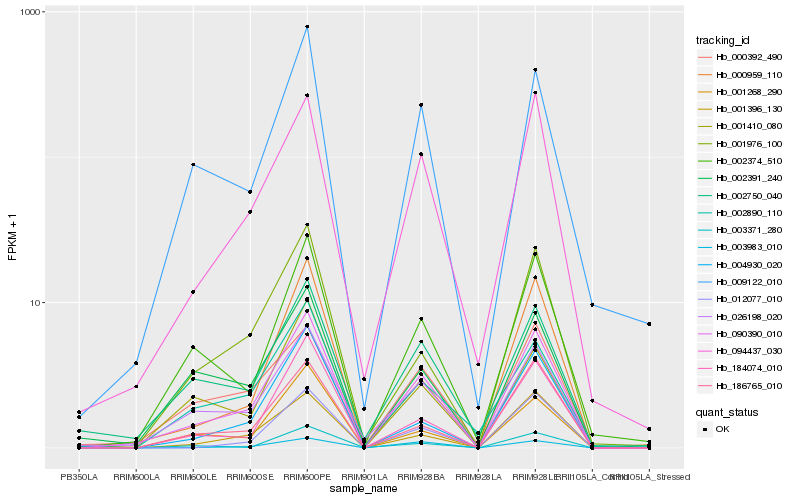
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_090390_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s28850g [Populus trichocarpa] |
| 2 |
Hb_026198_020 |
0.1078302371 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 3 |
Hb_094437_030 |
0.1090132504 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 4 |
Hb_002374_510 |
0.1228812553 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 5 |
Hb_009122_010 |
0.1233256517 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_001396_130 |
0.130773315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003983_010 |
0.1345865394 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 8 |
Hb_184074_010 |
0.1367506359 |
- |
- |
PREDICTED: uncharacterized protein LOC105639054 [Jatropha curcas] |
| 9 |
Hb_001976_100 |
0.1369923752 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_186765_010 |
0.1385643559 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 11 |
Hb_002750_040 |
0.1390981129 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 12 |
Hb_000392_490 |
0.1410826936 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003371_280 |
0.1418403923 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_012077_010 |
0.1443064024 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 15 |
Hb_002391_240 |
0.1445297758 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000959_110 |
0.1465355085 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 17 |
Hb_002890_110 |
0.1489195331 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 18 |
Hb_001268_290 |
0.1490906556 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 19 |
Hb_004930_020 |
0.1502544992 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 20 |
Hb_001410_080 |
0.1520331902 |
- |
- |
ring finger protein, putative [Ricinus communis] |