| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
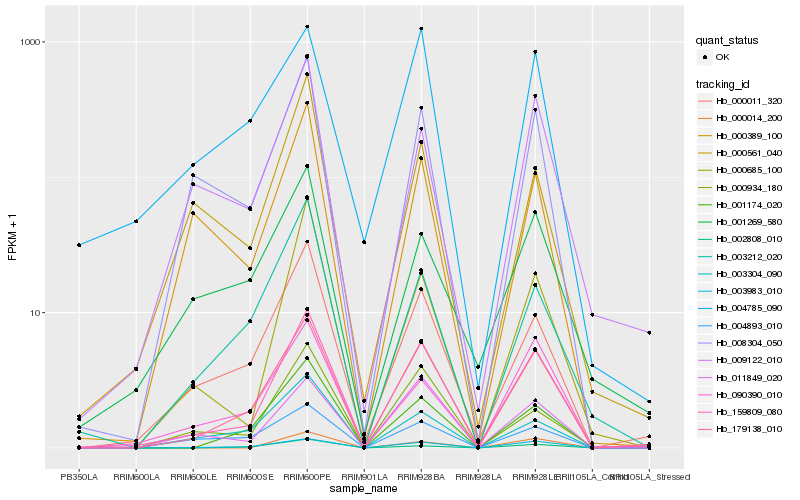
Hb_159809_080 |
0.0 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 2 |
Hb_179138_010 |
0.0842539782 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 3 |
Hb_003983_010 |
0.1166254565 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 4 |
Hb_000011_320 |
0.1239875981 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 5 |
Hb_008304_050 |
0.1406685384 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 6 |
Hb_001174_020 |
0.1486419901 |
- |
- |
- |
| 7 |
Hb_011849_020 |
0.1563827295 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |
| 8 |
Hb_009122_010 |
0.159251735 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000685_100 |
0.1600799873 |
- |
- |
hypothetical protein POPTR_0001s10630g [Populus trichocarpa] |
| 10 |
Hb_000934_180 |
0.1663407128 |
- |
- |
hypothetical protein JCGZ_25237 [Jatropha curcas] |
| 11 |
Hb_000389_100 |
0.1677310434 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 12 |
Hb_000014_200 |
0.1713872587 |
- |
- |
PREDICTED: uncharacterized protein LOC105775355 [Gossypium raimondii] |
| 13 |
Hb_090390_010 |
0.1756317259 |
- |
- |
hypothetical protein POPTR_0006s28850g [Populus trichocarpa] |
| 14 |
Hb_004893_010 |
0.176395906 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 15 |
Hb_004785_090 |
0.1782448283 |
- |
- |
hypothetical protein MIMGU_mgv1a018797mg [Erythranthe guttata] |
| 16 |
Hb_003212_020 |
0.1788625405 |
- |
- |
hypothetical protein PHAVU_009G114800g [Phaseolus vulgaris] |
| 17 |
Hb_001269_580 |
0.179638144 |
- |
- |
PREDICTED: shikimate O-hydroxycinnamoyltransferase [Jatropha curcas] |
| 18 |
Hb_003304_090 |
0.1804281211 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 19 |
Hb_000561_040 |
0.1828713746 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 20 |
Hb_002808_010 |
0.1836638802 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |