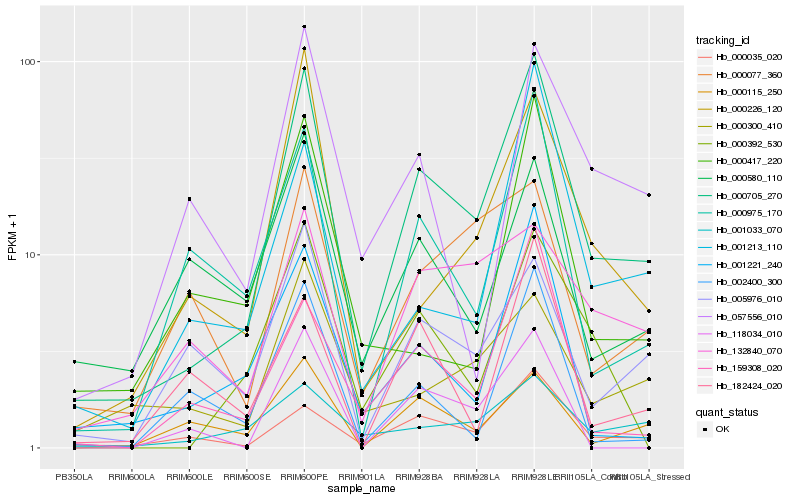
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000705_270 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105131117 [Populus euphratica] |
| 2 |
Hb_000035_020 |
0.1834180274 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 3 |
Hb_000975_170 |
0.1977119578 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001221_240 |
0.2033504639 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000392_530 |
0.2075061305 |
- |
- |
hypothetical protein PHAVU_003G0695001g, partial [Phaseolus vulgaris] |
| 6 |
Hb_159308_020 |
0.2129460798 |
- |
- |
hypothetical protein B456_002G027900 [Gossypium raimondii] |
| 7 |
Hb_000226_120 |
0.213299664 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005976_010 |
0.2136989832 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001213_110 |
0.2185312486 |
- |
- |
SOUL heme-binding family protein [Populus trichocarpa] |
| 10 |
Hb_001033_070 |
0.2194529841 |
- |
- |
seryl-tRNA synthetase, putative [Ricinus communis] |
| 11 |
Hb_057556_010 |
0.2213833335 |
- |
- |
Light-regulated protein precursor, putative [Ricinus communis] |
| 12 |
Hb_000115_250 |
0.2273601184 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 13 |
Hb_002400_300 |
0.230840822 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 14 |
Hb_000300_410 |
0.2309287931 |
- |
- |
hypothetical protein POPTR_0013s05860g [Populus trichocarpa] |
| 15 |
Hb_118034_010 |
0.2316668846 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 16 |
Hb_000580_110 |
0.2356270248 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 17 |
Hb_132840_070 |
0.2368274501 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 18 |
Hb_182424_020 |
0.2376439934 |
- |
- |
hypothetical protein POPTR_0006s24190g [Populus trichocarpa] |
| 19 |
Hb_000417_220 |
0.23893513 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Cucumis melo] |
| 20 |
Hb_000077_360 |
0.2391780406 |
- |
- |
sugar transporter, putative [Ricinus communis] |